It's great to see the prize go to scientists who are under 50, recognizing work that was done less than a decade ago. When the prize recognizes very old work too often, the Nobel loses much of its excitement and risks losing its relevance to current science. The original Nature paper by Fire, Mello, and their colleagues, came out in Feb. 1998. Since that time, RNAi has become huge in the field of biology - both in terms of its role in the cell, as a powerful genetics tool, and as a potential way to do gene therapy.
RNAi, or RNA interference, is when the expression of a single gene is shut off by the presence of foreign RNA molecules that match part of the sequence of that gene. RNAi occurs in plants, animals, and fungi, and most likely evolved as a defense against viruses and other genomic parasites (such as transposons - they're like viruses that spread through the genome and never leave the cell; we all have a lot of these parasites hanging around in our genomes).
What did Fire and Mello do? A look at their work illustrates how major scientific discoveries don't just come out of thin air - they are based on the groundwork usually laid down by several different research groups. Fire and Mello didn't discover RNAi - scientists had known for years that injecting small RNA molecules into organisms like round worms (such as C. elegans - the organism Fire and Mello work with) could inhibit the expression of single genes. Similar phenomena had been described for both plants and bread mold. (Bread mold - N. crassa - is another weird organism that has been used to study many basic cellular processes.)
People hypothesized that it worked because injected 'anti-sense' RNA bound (or 'hybridized') to the cell's messenger RNA (mRNA) and prevented that mRNA from serving as a template for synthesizing new protein:
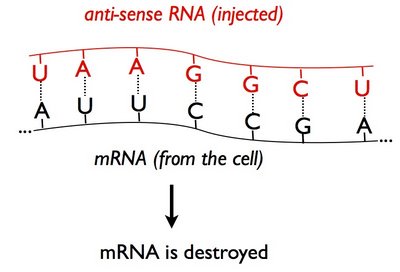
(Recall that the letters A,C,G, and in RNA, U stand for chemical groups that match up with each other - A pairs with U, C pairs with G.)
But there were problems with this idea - it also worked with 'sense' RNA as well, which would not bind to the mRNA molecule. Also, a small number of injected RNA molecules, not nearly enough to bind up all of the mRNA produced by a partuclar gene, could cause RNAi - suggesting that the injected RNA molecules were reused over and over to break down the mRNA (that is, they funtioned catalytically). This effect could even been seen in the next generation of organisms, that had not been injected with foreign RNA.
Fire, Mello and colleagues came up with the brilliant idea that the phenomenon might be caused by double-stranded RNA molecules, which would have been present as contaminants in any injected sample of RNA. Such double-stranded molecules would not work by simply hybridizing to the mRNA (as shown in the figure above). The process would have to work by some other mechanism, which at the time was completely unknown.
So Fire and Mello deliberately prepared sense, anti-sense, and double-stranded RNAs, injected them into worms, and looked to see which RNA molecules would be most effective at knocking down gene expression.
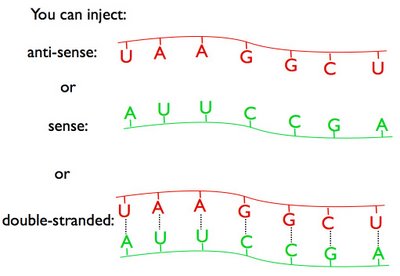
They picked a clever target gene to knock down - unc-22, a gene coding for a muscle protein. When that muscle protein is absent, the worms uncontrollably twitch. So Fire and Mello injected the different RNA molecules into worm embryos, and looked to see which ones resulted in twitching adult worms, indicating that the unc-22 gene had been shut off. Their results conclusively demonstrated that it was the double-stranded RNA molecules, not the individual sense or anti-sense molecules, that caused RNAi.
This paved the way for later discoveries figuring out excatly how this process works, and for making huge collections of these RNAs that can be used to knock down just about any gene you want in model organisms like flies and worms. Until RNAi, you couldn't shut off genes so easily, except in microorganisms like yeast (which is one reason why yeast is so useful for studying basic molecular biology). Now, you can do those things in multi-cellular organisms, and hardly a week goes by without the report of some new discovery made using RNAi. These double-stranded RNA molecules are even being tested as drugs in humans, to shut off aberrant genes in people with certain diseases. This was certainly a timely and well-deserved Nobel prize.




1 comment:
Thanks for the link. It's fascinating how this process is widespread among diverse eukaryotes.
Post a Comment