The NY Times is reporting on a conference at Rice University about women and science. (Free registration required.)
I've probably already said more than I should have about this subject, but there is stuff in this article that I can't let pass without comment.
I do agree that:
- Women make great scientists. We should have more of them.
- Women have faced severe discrimination on the road to a successful science career, especially the generation that currently holds most tenured faculty positions.
I think the greatest barrier women face in science today is not men looking down on them - it's the disproportionate cost borne by women in a career that demands severe sacrifices of people who are trying to juggle work and family. I believe that in academia there are fewer mechanisms of instutional support for women in tenure-track jobs who need time for family than there are in business or other professions like law or medicine. In terms of managing the grant funding for a research lab and training graduate students, women who hope to get tenure but take time off when their kids are young are at a severe disadvantage. I think we need to fix this (like 'start-up funds' or reserved grants for women who need to get the lab going again after maternity leave), so that the profession does not lose these talented women scientists.
What gets me riled up is the kind of garbage you can find in the Times article today - the claim that women will be judged more harshly than men if they don't use the correct balance of 'we' and 'I' when giving a talk on the collective work in their labs, that women can't be as aggressive as men when negotiating for resources, that they are excluded from networking, and that women are widely viewed by their male professional colleagues as "unsuited" for science. This is simply not true of the colleagues I work with.
One of the more egregious claims reported in the Times is that men show up to faculty job talks in jeans while women have to come in a suit. I have been to a lot of these talks - nobody who is serious about getting the job shows up in jeans, male or female. I have no clue where that ridiculous claim came from. Scientists, men and women, are notorious for dressing casual around the lab, but not when they are applying for competitive tenure-track jobs.
I can't speak for fields outside of my own - research in the basic life sciences, typically done in basic science departments at medical schools. Maybe it's different in all the other fields - social sciences, clinical sciences, engineering. Maybe it's a generational thing - I was born too late (or I'm too male) to really be attuned to the persistent prevalence of sexist attitudes in science. But I really don't see science as a major bastion of misogyny (which yes, it once was). The scientists I know are socially progressive and usually quite politically liberal. I just can't agree that the major reason for the attrition of women in science is the attitude of their male colleagues. The problems are structural problems in the career path which take a high toll on women who chose to have children at some point in their careers.
Sunday, December 31, 2006
Are some gene names too demeaning?
Recently, science journals have reported on criticism of whimsical gene names - critics say that names like sonic hedgehog and lunatic fringe are offensive to people (or parents of children) who have a disease caused by an altered variant of one of these genes. This is also occasionally being picked up by the larger media - I heard a brief reference to this debate on NPR the other day. In the Nov. 9 issue of Nature (yes, I know I'm way behind on my news) we find this news brief (subscription required). It says:
"A survey by the Gene Nomenclature Committee of the Human Genome Organisation, based at University College London, came up with ten genes that have "inappropriate, demeaning and pejorative" names, many of which are linked to eponymous developmental defects."
I agree that we shouldn't tell parents that their child has a bad sonic hedgehog gene, but the above sentence is wrong on so many levels. The gene names may be inappropriate, but, considering that these names came from the fly versions of the genes (since these genes were first discovered in flies), it makes no sense to call them "demeaning and pejorative." Nor does it make any sense to talk about the "eponymous developmental defects" since the genes are named after the fly develpmental defects, not the human ones. (Humans with mutations in one of the hedgehog genes do not look like hedgehogs, but fly embryos, with some creative imagination, do.)
What bugs me about this stupid debate is the scolding tone taken by the critics. Fly geneticists shouldn't give a damn about curbing the craziness of their gene names just because somewhere down the line the same gene may be involved in human disease. Science is a career that involves passion and fun, and biologists shouldn't walk around being excessively somber about their research, just because it may have implications for human disease.
The solution to this dilemma is simple, and shouldn't require the lecturing of the Orwellian sounding Gene Nomenclature Committee of the Human Genome Organisation. (I confess I have no clue who these people are.) Sonic Hedgehog and other such genes are frequently referred to by their abbreviations, such as Shh. This is how physicians should refer to them when speaking with patients or writing for clinical journals. Physicians can explain to their patients, who are often curious and will go do a literature search on their disease, that the gene Shh is named after the fly gene sonic hedgehog, which was named that way because it is somehow related to the hedgehog gene, which was named that way because mutant embryos look like hedgehogs. It's that simple, and nobody should be offended. And those of us who work on model organisms should be as whimsical as we want to be.
"A survey by the Gene Nomenclature Committee of the Human Genome Organisation, based at University College London, came up with ten genes that have "inappropriate, demeaning and pejorative" names, many of which are linked to eponymous developmental defects."
I agree that we shouldn't tell parents that their child has a bad sonic hedgehog gene, but the above sentence is wrong on so many levels. The gene names may be inappropriate, but, considering that these names came from the fly versions of the genes (since these genes were first discovered in flies), it makes no sense to call them "demeaning and pejorative." Nor does it make any sense to talk about the "eponymous developmental defects" since the genes are named after the fly develpmental defects, not the human ones. (Humans with mutations in one of the hedgehog genes do not look like hedgehogs, but fly embryos, with some creative imagination, do.)
What bugs me about this stupid debate is the scolding tone taken by the critics. Fly geneticists shouldn't give a damn about curbing the craziness of their gene names just because somewhere down the line the same gene may be involved in human disease. Science is a career that involves passion and fun, and biologists shouldn't walk around being excessively somber about their research, just because it may have implications for human disease.
The solution to this dilemma is simple, and shouldn't require the lecturing of the Orwellian sounding Gene Nomenclature Committee of the Human Genome Organisation. (I confess I have no clue who these people are.) Sonic Hedgehog and other such genes are frequently referred to by their abbreviations, such as Shh. This is how physicians should refer to them when speaking with patients or writing for clinical journals. Physicians can explain to their patients, who are often curious and will go do a literature search on their disease, that the gene Shh is named after the fly gene sonic hedgehog, which was named that way because it is somehow related to the hedgehog gene, which was named that way because mutant embryos look like hedgehogs. It's that simple, and nobody should be offended. And those of us who work on model organisms should be as whimsical as we want to be.
Saturday, December 09, 2006
A Scientist's take on Thomas Pynchon's Against the Day
Thomas Pynchon is an author that scientists should love. No other first-rate novelist has engaged more deeply with the actual content of our civilization's scientific discoveries, especially math and physics. His latest book, Against The Day, is no exception, and in fact incorporates even more scientific content than the rocketry-obsessed Gravity's Rainbow. This is because Against the Day spans the scientifically rich years around the start of the 20th century. Although a lot of my colleagues seem to have never heard of the guy, at least one physicist shares my taste for contemporary fiction and love for Pynchon's books.
What's great about the science in Pynchon's novels is that he knows his material well enough to purposely and very effectively blur the lines between what we'd call serious science and crank science. Ambiguity is a hallmark of Pynchon's writing, and the late (and not-so-late) 19th century advances in math and physics are fertile ground for Pynchon to plow: Grassman algebra, Maxwell's equations on electromagnetic theory, Hamilton's quarternions (there is even a crazy quarternion song at a Quarterionist party that begins: "O, the, Quizzical, queer Quarter-nioneer, that creature of i-j-k"!), Riemann surfaces, the Michelson-Morely experiment, etc. Throughout the book, Pynchon features fanatical crank devotees of various scientific ideas right alongside legitimate scientists. For example, various more or less nutty groups show up in Ohio for the Michelson-Morely experiment, which takes place early in the book.
Louis Menand, in a review of Against the Day for the New Yorker put it this way:
"We [today] know (roughly) how it all turned out, but if we had been living in those years it would have been impossible to sort out the fantastical possibilities from the plausible ones. Maybe we could split time and be in two places at once, or travel backward and forward at will, or maintain parallel lives in parallel universes."
Yet Pynchon is by no means anti-science. His books are about forces that shape and drive people's lives, and how people respond to them - to the personal, societal, and physical forces that leave us uncertain about what's true and what control we have over our own lives. Science and technology have been major forces acting on all of our lives, and Pynchon handles this theme better (and with more complexity and nuance) than any novelist I've come accross, including great science fiction novelists like Asimov and Dick.
This is why I think Laura Miller is seriously wrong, reviewing Against the Day for Salon (under the ridiculously portentous title "The Fall of the House of Pynchon") when she writes that younger writers now have a "grasp of the systems that fascinate Pynchon -- science, capitalism, religion, politics, technology -- [that] is surer, more nuanced, more adult and inevitably yields more insight into how those systems work than Pynchon offers here." While I love the writers Miller mentions (David Foster Wallace and Neal Stephenson), nobody can do what Pynchon does as well as he does it. Maybe you need to have some basic literacy in math and science (and the aid of Wikipedia or Penrose's The Road to Reality) to really appreciate it, and as many professional reviewers of Against the Day admitted, 7th grade math is enough to confuse most people. (Pynchon started out as a physics or engineering major at Cornell before swtiching to English, and he worked as a technical writer for Boeing, so he has some background. But for most of it, I imagine he has to be self-taught, something he obviously has a great capacity for.)
Which leads me on to the subject of the high-profile negative reviews the book has received. (If you're bored by literary dust-ups, here is a good place to stop reading this post!) There were several prominent, early negative reviews that overshadowed the many positive ones. Kakutani, in the NY Times, trashed the book, but it's not surprising - she has a long history of hating this kind of fiction, and I doubt she really spent the necessary time with the 1,100 page novel - her mind was made up before the book was even published. For scientists, it's like having the guy who is philosophically opposed to your work review your manuscript submission - that's the kind of guy you ask editors to exclude from lists of potential reviewers. Incidentally, the NY Times also published a longer, very positive review by someone else. (For a positive review, and a ton of links to other reviews, check out the Literary Saloon.)
The common theme in most of the negative reviews (and a common complaint made by critics of Pynchon in general) is that his characters are flat and devoid of humanity. I find Pynchon's books to be some of the most human books I've read. The best way to make this clear is by a comparison with modern art: Some people think that characters in a novel are inhuman unless they are fully fleshed out, filled with rich and undistorted detail that allows us to know all about them, much like a Delacroix painting (like this or this.) But in great modern art, artists can capture the essential humanity of a character in more sparse, angular strokes, without the flood of realistic detail - like Picasso here and here. If Pynchon's character's are cold and inhuman, than so are Picasso's.
If you're tempted to read Against the Day, don't let the high-profile negative reviews scare you away. Pynchon's books are complex, but in a good way - the way wine is complex compared with Coke, or a good coffee compared with Nesquick. His prose is stunning, and I, for one, can't get enough of it.
What's great about the science in Pynchon's novels is that he knows his material well enough to purposely and very effectively blur the lines between what we'd call serious science and crank science. Ambiguity is a hallmark of Pynchon's writing, and the late (and not-so-late) 19th century advances in math and physics are fertile ground for Pynchon to plow: Grassman algebra, Maxwell's equations on electromagnetic theory, Hamilton's quarternions (there is even a crazy quarternion song at a Quarterionist party that begins: "O, the, Quizzical, queer Quarter-nioneer, that creature of i-j-k"!), Riemann surfaces, the Michelson-Morely experiment, etc. Throughout the book, Pynchon features fanatical crank devotees of various scientific ideas right alongside legitimate scientists. For example, various more or less nutty groups show up in Ohio for the Michelson-Morely experiment, which takes place early in the book.
Louis Menand, in a review of Against the Day for the New Yorker put it this way:
"We [today] know (roughly) how it all turned out, but if we had been living in those years it would have been impossible to sort out the fantastical possibilities from the plausible ones. Maybe we could split time and be in two places at once, or travel backward and forward at will, or maintain parallel lives in parallel universes."
Yet Pynchon is by no means anti-science. His books are about forces that shape and drive people's lives, and how people respond to them - to the personal, societal, and physical forces that leave us uncertain about what's true and what control we have over our own lives. Science and technology have been major forces acting on all of our lives, and Pynchon handles this theme better (and with more complexity and nuance) than any novelist I've come accross, including great science fiction novelists like Asimov and Dick.
This is why I think Laura Miller is seriously wrong, reviewing Against the Day for Salon (under the ridiculously portentous title "The Fall of the House of Pynchon") when she writes that younger writers now have a "grasp of the systems that fascinate Pynchon -- science, capitalism, religion, politics, technology -- [that] is surer, more nuanced, more adult and inevitably yields more insight into how those systems work than Pynchon offers here." While I love the writers Miller mentions (David Foster Wallace and Neal Stephenson), nobody can do what Pynchon does as well as he does it. Maybe you need to have some basic literacy in math and science (and the aid of Wikipedia or Penrose's The Road to Reality) to really appreciate it, and as many professional reviewers of Against the Day admitted, 7th grade math is enough to confuse most people. (Pynchon started out as a physics or engineering major at Cornell before swtiching to English, and he worked as a technical writer for Boeing, so he has some background. But for most of it, I imagine he has to be self-taught, something he obviously has a great capacity for.)
Which leads me on to the subject of the high-profile negative reviews the book has received. (If you're bored by literary dust-ups, here is a good place to stop reading this post!) There were several prominent, early negative reviews that overshadowed the many positive ones. Kakutani, in the NY Times, trashed the book, but it's not surprising - she has a long history of hating this kind of fiction, and I doubt she really spent the necessary time with the 1,100 page novel - her mind was made up before the book was even published. For scientists, it's like having the guy who is philosophically opposed to your work review your manuscript submission - that's the kind of guy you ask editors to exclude from lists of potential reviewers. Incidentally, the NY Times also published a longer, very positive review by someone else. (For a positive review, and a ton of links to other reviews, check out the Literary Saloon.)
The common theme in most of the negative reviews (and a common complaint made by critics of Pynchon in general) is that his characters are flat and devoid of humanity. I find Pynchon's books to be some of the most human books I've read. The best way to make this clear is by a comparison with modern art: Some people think that characters in a novel are inhuman unless they are fully fleshed out, filled with rich and undistorted detail that allows us to know all about them, much like a Delacroix painting (like this or this.) But in great modern art, artists can capture the essential humanity of a character in more sparse, angular strokes, without the flood of realistic detail - like Picasso here and here. If Pynchon's character's are cold and inhuman, than so are Picasso's.
If you're tempted to read Against the Day, don't let the high-profile negative reviews scare you away. Pynchon's books are complex, but in a good way - the way wine is complex compared with Coke, or a good coffee compared with Nesquick. His prose is stunning, and I, for one, can't get enough of it.
Thursday, December 07, 2006
Going though the NIH review process
One of the most critical things a postdoc in science has to do is get independent funding. Even if your mentor is rich, proving that you can put together a research proposal and successfully obtain funding is a critical career step.
I submitted four funding applications this year, my first year as a postdoc. It's a long, slow process - each funding application has different length and content requirements - ranging from 10 pages not counting references, to 5 including references. By far I found the NIH applicatio to be the most onerous. (The NIH was the only US government agency I applied to - the rest were private foundations.) The application includes a massive, 104 page instuction manual for filling out all the damn forms. And then it takes months and months to go through the whole cycle. I submitted my application in August, and I just got my reviews back. I won't know about a final funding decision until February at the latest.
It was an interesting experience getting these first reviews. NIH fellowship applications are scored on a scale of 100-500, with 100 being the best. (Actually, I don't think they really score proposals that earn a score worse than maybe 300 - the applications considered to be in the worst 40% are 'triaged' and not reviewed further.) Applicants also get a summary of the discussion that took place in the review session, and written critiques from three reviewers.
Each of my three reviewers disagrees with the other two at some point. It's amusing to read some of the examples:
Reviewer 1: "The application is for an average candidate in an outstanding environment..."
Reviewer 3: "This is a very strong application from an outstanding candidate..."
Reviewer 1: "However, there is little discussion of the possible outcomes of each set of experiments, or the possible pitfalls and stumbling blocks that might be encountered if all does not go as planned..."
Reviewer 2: "This specific aim was concise, comprehensive and well-written. It
addresses potential concerns..."
As long and cumbersome as the overall process is, I got fair, good-faith reviews from all the people involved, even though I disagree with some points here and there. My program officer at the NIH (the person handling my application) said I was in the 'maybe' category. The worst part is that I have to wait a few more months (by which time I will have completed my first year as a postdoc) to hear the final decision. And then I may have to do a minor rewrite, and go through the whole damn cycle again. It's a long process for something that's supposed to happen during a relatively short postdoctoral period.
Now that I'm obviously much wiser about this, having gone through the whole process exactly 0.75 times, I have some advice for any grad students who stumble across this blog:
- find a mentor who has excellent grant writing skills and get him or her to give you a ton of advice
- find a postdoc who already has fellowship funding, and get a copy of that proposal. Reading someone else's successful proposal is invaluable.
- keep sending in applications everywhere you can until someone gives you money.
I submitted four funding applications this year, my first year as a postdoc. It's a long, slow process - each funding application has different length and content requirements - ranging from 10 pages not counting references, to 5 including references. By far I found the NIH applicatio to be the most onerous. (The NIH was the only US government agency I applied to - the rest were private foundations.) The application includes a massive, 104 page instuction manual for filling out all the damn forms. And then it takes months and months to go through the whole cycle. I submitted my application in August, and I just got my reviews back. I won't know about a final funding decision until February at the latest.
It was an interesting experience getting these first reviews. NIH fellowship applications are scored on a scale of 100-500, with 100 being the best. (Actually, I don't think they really score proposals that earn a score worse than maybe 300 - the applications considered to be in the worst 40% are 'triaged' and not reviewed further.) Applicants also get a summary of the discussion that took place in the review session, and written critiques from three reviewers.
Each of my three reviewers disagrees with the other two at some point. It's amusing to read some of the examples:
Reviewer 1: "The application is for an average candidate in an outstanding environment..."
Reviewer 3: "This is a very strong application from an outstanding candidate..."
Reviewer 1: "However, there is little discussion of the possible outcomes of each set of experiments, or the possible pitfalls and stumbling blocks that might be encountered if all does not go as planned..."
Reviewer 2: "This specific aim was concise, comprehensive and well-written. It
addresses potential concerns..."
As long and cumbersome as the overall process is, I got fair, good-faith reviews from all the people involved, even though I disagree with some points here and there. My program officer at the NIH (the person handling my application) said I was in the 'maybe' category. The worst part is that I have to wait a few more months (by which time I will have completed my first year as a postdoc) to hear the final decision. And then I may have to do a minor rewrite, and go through the whole damn cycle again. It's a long process for something that's supposed to happen during a relatively short postdoctoral period.
Now that I'm obviously much wiser about this, having gone through the whole process exactly 0.75 times, I have some advice for any grad students who stumble across this blog:
- find a mentor who has excellent grant writing skills and get him or her to give you a ton of advice
- find a postdoc who already has fellowship funding, and get a copy of that proposal. Reading someone else's successful proposal is invaluable.
- keep sending in applications everywhere you can until someone gives you money.
Wednesday, December 06, 2006
Molecular biologists don't understand molecular biology, but Creationist engineers do?
Via Pharyngula I heard about these thoughts by a Creationist engineer musing on the contents of a molecular biology textbook (the link is to All-Too-Common Dissent, a pro science blog written by a scientist, where you can find the link to the original Creationist post):
"My hypothesis is that the field of molecular biology is simply not understood by the majority of biologists and thus pretty secure from rational debate by laymen. By claiming that this discipline (which they probably don't understand either) proves Darwinism and that Darwinism is vital to understanding molecular biology, the Creationists can be silenced, humiliated and put in their place by simply invoking superior knowledge. More malpractice?"
And there's this gem:
"It [this molecular biology textbook the guy was looking at] has a considerable Physical Chemistry and Organic Chemistry component which would make it intimidating for the large majority of biologists, but this subject is really foundational to understanding the molecular foundations of genetics"
Eh? The field of molecular biology is not understood by the people who are actually doing professional research on the subject, but it is understood by a fundamentalist Christian engineer?
So biologists don't know enough math, chemistry and physics to understand what they're doing? I hear comments like this every so often, like the guy who told me that his chemist friend doesn't consider biochemists real chemists. Another example is the first comment to the All-To-Common Dissent post where one guy says this:
"I'm an engineer but also did biology at university for one year. Let me say that the math and physics done at the university level by biology students nowhere approaches real math or physics; it's barely beyond grade 12 level. That said, the only biology you get in engineering is a few things about bacteria when it came to municipal water engineering, so not a whole lot, and certainly less than what biologists got in math and physics."
One year of freshman biology does not really give you an idea of the kind of quantitative training biologists get. I definitely think that biologists these days need more quantitative training to deal with some of today's most exciting research questions (and the educational trend is going in that direction). However, as Scott showed in the original post on All-To-Common Dissent, undergraduate biology curricula require plenty of math, physics, and chemistry - enough to truly grasp molecular biology as well as it's understood today, and usually much more chemistry than engineers are required to learn (chemical engineers excepted of course).
Biochemists get serious chemistry training - as an undergraduate, I had 5 semesters of chemistry (plus labs) before I took a year's worth of biochemistry; I also had 4 semesters of physics, including a class on quantum mechanics where we practiced solving the Schroedinger equation (admittedly just the easier time-independent form). Most biochemistry programs require 1-2 semsters of physical chemistry as well (I didn't major in biochem, so I didn't take p-chem). Anyone who has taken an upper-level undergrad course in physiology can vouch for the quantitative nature of that field. (The course I took involved more math than was in my chemistry classes.)
It's true that biologists generally don't go beyond two semesters of calculus and maybe introductory linear algebra, but frankly you don't need more than that to be a successful molecular biologist who understands the field. (Also, most of us learn much of our math in physics and chemistry courses, not math courses.)
At the graduate level, almost all programs require anyone in biology to take a graduate-level biochemistry course, which involves a lot of homework problems dealing with kinetics and thermodynamics (physical chemistry) and enzyme reaction mechanisms (electron-pushing organic chemistry) - maybe intimidating, as the Creationist engineer claims, but the fact is, all of us biology PhDs had to take and pass it.
And I'm not even getting into genetics, which has its own quantitative foundation.
Engineers and physicists like to pretend that biologists stop their quantitative training as soon as they finish high school. As we've seen, that's not true, but there is another important point - nobody, not even engineers and physicists, has really figured out (yet - I'm still hopeful) how to effectively, compellingly, use more sophisticated math to produce a deeper understanding of biology. Computer scientists and statisticians have contributed a lot to genome sequence analysis, but we have not yet produced very predictive theories or models of cellular behavior that are rooted in the physical behavior of the components of the cell. That may require more math than what biologists are using now. However I doubt that biology (or engineering, for that matter) will ever require the kind of math used by today's theoretical physicists. For the time being, most biologsts know enough math and chemistry to understand the discipline they work in.
"My hypothesis is that the field of molecular biology is simply not understood by the majority of biologists and thus pretty secure from rational debate by laymen. By claiming that this discipline (which they probably don't understand either) proves Darwinism and that Darwinism is vital to understanding molecular biology, the Creationists can be silenced, humiliated and put in their place by simply invoking superior knowledge. More malpractice?"
And there's this gem:
"It [this molecular biology textbook the guy was looking at] has a considerable Physical Chemistry and Organic Chemistry component which would make it intimidating for the large majority of biologists, but this subject is really foundational to understanding the molecular foundations of genetics"
Eh? The field of molecular biology is not understood by the people who are actually doing professional research on the subject, but it is understood by a fundamentalist Christian engineer?
So biologists don't know enough math, chemistry and physics to understand what they're doing? I hear comments like this every so often, like the guy who told me that his chemist friend doesn't consider biochemists real chemists. Another example is the first comment to the All-To-Common Dissent post where one guy says this:
"I'm an engineer but also did biology at university for one year. Let me say that the math and physics done at the university level by biology students nowhere approaches real math or physics; it's barely beyond grade 12 level. That said, the only biology you get in engineering is a few things about bacteria when it came to municipal water engineering, so not a whole lot, and certainly less than what biologists got in math and physics."
One year of freshman biology does not really give you an idea of the kind of quantitative training biologists get. I definitely think that biologists these days need more quantitative training to deal with some of today's most exciting research questions (and the educational trend is going in that direction). However, as Scott showed in the original post on All-To-Common Dissent, undergraduate biology curricula require plenty of math, physics, and chemistry - enough to truly grasp molecular biology as well as it's understood today, and usually much more chemistry than engineers are required to learn (chemical engineers excepted of course).
Biochemists get serious chemistry training - as an undergraduate, I had 5 semesters of chemistry (plus labs) before I took a year's worth of biochemistry; I also had 4 semesters of physics, including a class on quantum mechanics where we practiced solving the Schroedinger equation (admittedly just the easier time-independent form). Most biochemistry programs require 1-2 semsters of physical chemistry as well (I didn't major in biochem, so I didn't take p-chem). Anyone who has taken an upper-level undergrad course in physiology can vouch for the quantitative nature of that field. (The course I took involved more math than was in my chemistry classes.)
It's true that biologists generally don't go beyond two semesters of calculus and maybe introductory linear algebra, but frankly you don't need more than that to be a successful molecular biologist who understands the field. (Also, most of us learn much of our math in physics and chemistry courses, not math courses.)
At the graduate level, almost all programs require anyone in biology to take a graduate-level biochemistry course, which involves a lot of homework problems dealing with kinetics and thermodynamics (physical chemistry) and enzyme reaction mechanisms (electron-pushing organic chemistry) - maybe intimidating, as the Creationist engineer claims, but the fact is, all of us biology PhDs had to take and pass it.
And I'm not even getting into genetics, which has its own quantitative foundation.
Engineers and physicists like to pretend that biologists stop their quantitative training as soon as they finish high school. As we've seen, that's not true, but there is another important point - nobody, not even engineers and physicists, has really figured out (yet - I'm still hopeful) how to effectively, compellingly, use more sophisticated math to produce a deeper understanding of biology. Computer scientists and statisticians have contributed a lot to genome sequence analysis, but we have not yet produced very predictive theories or models of cellular behavior that are rooted in the physical behavior of the components of the cell. That may require more math than what biologists are using now. However I doubt that biology (or engineering, for that matter) will ever require the kind of math used by today's theoretical physicists. For the time being, most biologsts know enough math and chemistry to understand the discipline they work in.
Sunday, December 03, 2006
Best Science Books
Once more, it's the most commercial time of the year, and every one is coming out with their Holiday Book recommendations. While the NY Times and the Washington Post put out the most well known best books list (and I'm peeved, but not surprised, that Thomas Pynchon and Richard Powers didn't make these top 10 lists), Discover Magazine has put out a list of the best 25 science books of all time.
The blog for the National Book Critics Circle notes that this list "is more about scientific umph than readability." Frankly, I don't even think that's true - I really can't see what bizarre criteria would generate a list that includes Newton's Principia (not that readable), Lovelock's Gaia (not much scientific umph), and Watson's Double Helix (very readable, but not a scientific milestone like the Principia). "Scientific umph" is frankly a bad criterion for a list of great scientific books, because major accomplishments in science are generally not first presented in book form (with a few obvious and older exceptions - Newton's Principia, Darwin's Origin. Sheer literary value is not a good criterion either, because you can have well-written 'science' books where the science itself is not really that substantial, but the writing is so good they're worth reading anyway. (Some people are going to hate me for this, but I find Lewis Thomas's books to be in this category. Personally, I don't find them as insightful about science as the books on my list below, but I know others disagree.)
A list of great books should have both scientific umph, and literary value, but in this sense: they should be compelling, well-written books that convey something deep about science - how it works, how its practitioners think, or about a specific concept. They should make a serious contribution to how we (scientists and the public) think about some aspect of science.
So I've put together my (currently partial) list of what I think are the best science books (in English), books which should be readable (though not necessarily easily readable - understanding science does take effort) by people who aren't professionals in the subject dealt with:
The Making of the Atomic Bomb, Richard Rhodes (1986) (and the 'sequel', Dark Sun, about the hydrogen bomb). This is the most amazing scientific history I've every read. Rhodes writes in absolutely stunning prose, and gives the best current historical account of the development of nuclear physics. Along with the elements of scientific history and biography, Rhodes also develops other themes, like the relationship of science to war, the state, and secrecy. Science writing doesn't get any better than this.
The Character of Physical Law, Richard Feynman (1965). I have fantasies about some day teaching an Intro to Science course using these incredible lectures about scientific reasoning. Feynman was a master lecturer (keep in mind that essentially all of Feynman's books are edited transcriptions of lectures). So while the prose, in a literary sense, may not be Pulitzer material, as lectures, Feynman's language is superb. This book provides very deep insight into scientific thinking, with (naturally) a large slant towards physics.
On the Origin of Species, Charles Dawrin (1859) (Also, The Descent of Man, which I think is a better second Darwin book to read than the Voyage of the Beagle.) One of the few books of the last 200 years to be in itself a major scientific contribution. The Origin is still popular because it is one of the most readable original scientific contributions out there. Darwin was apologetic about his writing style, but hey, in general those Victorians were pretty damn literate, so it's a good read. Actually, Darwin's style is quite good - he usually avoids the pathologically florid style which other scientific writing of that era often succumbed to. This book is substantial because it lays out a major scientific discovery and is an excellent example of scientific reasoning.
Feynman Lectures on Physics, Feynman, Leighton, and Sands (1964) (In principle, this should be generally readable, since it begins with Freshman physics; in reality, it's very tough. Lay readers can do almost as well with the subset of the lectures contained in 6 Easy Pieces and 6 Not So Easy Pieces.) Why am I giving this a separate entry, when I've already included The Character of Physical law? These lectures, while providing similar insight into general scientific reasoning, give an excellent presentation of specific scientific content - of what we have actually learned through scientific reasoning. If you're a person whose willing to plow through dense books by people like Kant, Wittgenstein, or James Joyce, the same effort with the lectures on physics will be well worth the payoff. In terms of language, again keep in mind that these are edited lectures - from this perspective, the language is very well crafted.
The Double Helix, James Waton (1968) This is a well-written account of one of the major discoveries in biology.
The Selfish Gene, Richard Dawkins (1976) This is sort of a popular book, sort of an original contribution, but like Darwin's Origin, a well-written exposition of one way of reasoning using evolutionary concepts.
What is Life, Erwin Schroedinger (1944) This is somewhat dated, and some of the ideas presented have been rejected. However, this book single handedly pushed many physicists into biology, some of whom turned out be major figures in the development of molecular biology. This book is valuable for more than just historical interest though - Schroedinger raises unsolved issues about the Origins of Life.
Genius, James Gleick (1992) (Chaos is another great Gleick book that I think is at the same level.) This is my favorite scientific biography of all time. Gleick, like Rhodes, is a superb writer. I almost didn't put this book on this list though, because I don't think Gleick effectively conveys the content of Feynman's scientific work. (In Gleick's defense, quantum electrodynamics is incredibly abstract and tough to convey.) But, as a work about what makes a scientist tick, and how a great scientist develops, this book ranks among the best.
Goedel, Escher, Bach, Douglas Hofstadter (1979) A great big, substantial meditation on math, music, and logic, composed in vivid, well-crafted language.
A Brief History of Time, Stephen Hawking (1988) A classic discussion of some of the big questions facing physics.
I'm going to stop the list here (out of sheer laziness, and to leave room for other great authors I haven't read yet), but obviously there are many more that can be included. There are a bunch of other books that are some of my personal favorites, but don't, for one reason or another, quite have the timeless/universal quality of the books above:
The Road To Reality, Guns, Germs, and Steel, Ahead of the Curve, The Demon-Haunted World, Human Natures, The Triumph and Tragedy of J. Robert Oppenheimer, The Creationists (this is a really well-written book about people trying to cope with their faith and the progress of modern biology), This is Biology, Boltzmann's Atom, Silent Spring... - this list can go on for awhile.
My favorite science book published in 2006 was Nicolas Wade's Before the Dawn, about human evolution. Wade is a skilled writer who can effectively convey scientific content in accessible language. On the downside, I don't think this book does enough to distinguish hypotheses and speculation from widely accepted scientific conclusions, always a tough issue when you're describing science at the frontier of a given field.
There's a lot of good stuff to read out there.
UPDATE 12-4-06: I forgot to include one major book that should have been on Discover's list: The Eight Day of Creation, by Horace Freeland Judson (1979) This is one of the most insightful histories of science. It deals with the molecular biology revolution, and is full of excellent material about how the founding molecular biologists chose their questions, thought about them, and largely solved them. There is no better book on molecular biology.
The blog for the National Book Critics Circle notes that this list "is more about scientific umph than readability." Frankly, I don't even think that's true - I really can't see what bizarre criteria would generate a list that includes Newton's Principia (not that readable), Lovelock's Gaia (not much scientific umph), and Watson's Double Helix (very readable, but not a scientific milestone like the Principia). "Scientific umph" is frankly a bad criterion for a list of great scientific books, because major accomplishments in science are generally not first presented in book form (with a few obvious and older exceptions - Newton's Principia, Darwin's Origin. Sheer literary value is not a good criterion either, because you can have well-written 'science' books where the science itself is not really that substantial, but the writing is so good they're worth reading anyway. (Some people are going to hate me for this, but I find Lewis Thomas's books to be in this category. Personally, I don't find them as insightful about science as the books on my list below, but I know others disagree.)
A list of great books should have both scientific umph, and literary value, but in this sense: they should be compelling, well-written books that convey something deep about science - how it works, how its practitioners think, or about a specific concept. They should make a serious contribution to how we (scientists and the public) think about some aspect of science.
So I've put together my (currently partial) list of what I think are the best science books (in English), books which should be readable (though not necessarily easily readable - understanding science does take effort) by people who aren't professionals in the subject dealt with:
The Making of the Atomic Bomb, Richard Rhodes (1986) (and the 'sequel', Dark Sun, about the hydrogen bomb). This is the most amazing scientific history I've every read. Rhodes writes in absolutely stunning prose, and gives the best current historical account of the development of nuclear physics. Along with the elements of scientific history and biography, Rhodes also develops other themes, like the relationship of science to war, the state, and secrecy. Science writing doesn't get any better than this.
The Character of Physical Law, Richard Feynman (1965). I have fantasies about some day teaching an Intro to Science course using these incredible lectures about scientific reasoning. Feynman was a master lecturer (keep in mind that essentially all of Feynman's books are edited transcriptions of lectures). So while the prose, in a literary sense, may not be Pulitzer material, as lectures, Feynman's language is superb. This book provides very deep insight into scientific thinking, with (naturally) a large slant towards physics.
On the Origin of Species, Charles Dawrin (1859) (Also, The Descent of Man, which I think is a better second Darwin book to read than the Voyage of the Beagle.) One of the few books of the last 200 years to be in itself a major scientific contribution. The Origin is still popular because it is one of the most readable original scientific contributions out there. Darwin was apologetic about his writing style, but hey, in general those Victorians were pretty damn literate, so it's a good read. Actually, Darwin's style is quite good - he usually avoids the pathologically florid style which other scientific writing of that era often succumbed to. This book is substantial because it lays out a major scientific discovery and is an excellent example of scientific reasoning.
Feynman Lectures on Physics, Feynman, Leighton, and Sands (1964) (In principle, this should be generally readable, since it begins with Freshman physics; in reality, it's very tough. Lay readers can do almost as well with the subset of the lectures contained in 6 Easy Pieces and 6 Not So Easy Pieces.) Why am I giving this a separate entry, when I've already included The Character of Physical law? These lectures, while providing similar insight into general scientific reasoning, give an excellent presentation of specific scientific content - of what we have actually learned through scientific reasoning. If you're a person whose willing to plow through dense books by people like Kant, Wittgenstein, or James Joyce, the same effort with the lectures on physics will be well worth the payoff. In terms of language, again keep in mind that these are edited lectures - from this perspective, the language is very well crafted.
The Double Helix, James Waton (1968) This is a well-written account of one of the major discoveries in biology.
The Selfish Gene, Richard Dawkins (1976) This is sort of a popular book, sort of an original contribution, but like Darwin's Origin, a well-written exposition of one way of reasoning using evolutionary concepts.
What is Life, Erwin Schroedinger (1944) This is somewhat dated, and some of the ideas presented have been rejected. However, this book single handedly pushed many physicists into biology, some of whom turned out be major figures in the development of molecular biology. This book is valuable for more than just historical interest though - Schroedinger raises unsolved issues about the Origins of Life.
Genius, James Gleick (1992) (Chaos is another great Gleick book that I think is at the same level.) This is my favorite scientific biography of all time. Gleick, like Rhodes, is a superb writer. I almost didn't put this book on this list though, because I don't think Gleick effectively conveys the content of Feynman's scientific work. (In Gleick's defense, quantum electrodynamics is incredibly abstract and tough to convey.) But, as a work about what makes a scientist tick, and how a great scientist develops, this book ranks among the best.
Goedel, Escher, Bach, Douglas Hofstadter (1979) A great big, substantial meditation on math, music, and logic, composed in vivid, well-crafted language.
A Brief History of Time, Stephen Hawking (1988) A classic discussion of some of the big questions facing physics.
I'm going to stop the list here (out of sheer laziness, and to leave room for other great authors I haven't read yet), but obviously there are many more that can be included. There are a bunch of other books that are some of my personal favorites, but don't, for one reason or another, quite have the timeless/universal quality of the books above:
The Road To Reality, Guns, Germs, and Steel, Ahead of the Curve, The Demon-Haunted World, Human Natures, The Triumph and Tragedy of J. Robert Oppenheimer, The Creationists (this is a really well-written book about people trying to cope with their faith and the progress of modern biology), This is Biology, Boltzmann's Atom, Silent Spring... - this list can go on for awhile.
My favorite science book published in 2006 was Nicolas Wade's Before the Dawn, about human evolution. Wade is a skilled writer who can effectively convey scientific content in accessible language. On the downside, I don't think this book does enough to distinguish hypotheses and speculation from widely accepted scientific conclusions, always a tough issue when you're describing science at the frontier of a given field.
There's a lot of good stuff to read out there.
UPDATE 12-4-06: I forgot to include one major book that should have been on Discover's list: The Eight Day of Creation, by Horace Freeland Judson (1979) This is one of the most insightful histories of science. It deals with the molecular biology revolution, and is full of excellent material about how the founding molecular biologists chose their questions, thought about them, and largely solved them. There is no better book on molecular biology.
Thursday, November 30, 2006
Sequencing technology and the Neanderthal Genome
You've probably read about the two recent papers in Science and Nature reporting the sequencing of portions of the Neanderthal Genome. (Subscription required for full text of the Nature and Science papers. Check out Google for the many news stories on this.) This is exciting work, but I'm not really going to comment about the signficance of the results - I think it's worth understanding how new sequencing technology enabled researchers to sequence 1 million bases of 38,000 year-old DNA.
Each group used a different sequencing technology, and as a result, their coverage of the genome differed widely: the Nature group got about 1 million base pairs of sequence, while the Science group got about 65,000 base pairs. (Recall that the human genome contains about 3 billion base pairs, and the Neanderthal genome was undoubtedly similar.) The Neanderthal genome is a challenge because, obviously, any DNA remaining in the 30,000-40,000 year old bones we have is highly fragmented, and the amount of contaminating DNA from microbes (not to mention scientists) is significant. To eventually cover the entire genome, we need a method that can generate lots and lots of sequence without being prohibitively expensive.
The Science group used a more traditional method, which works well for most large-scale genome sequencing efforts, but is not really well-suited for getting huge chunks of Neanderthal genome. As a result, this group obtained only 65,000 base pairs of DNA sequence (which is still a significant accomplishment.) The big problem with this method is that the fragments of DNA isolated from Neanderthal bones have to be cloned before they can be sequenced. (In layman's terms - the DNA fragments have to be placed inside circular pieces of DNA called plasmids, which can be then grown in large quantities inside bactieria.) Many fragments of Neanderthal DNA fail to be cloned at this point, meaning that you lose much of the sample that was painstakingly isolated from the ancient bones.
The Nature group used something called pyrosequencing, which is done on machines called 454 sequencers. Crucially, this technique does not require a cloning step, which means much more of the isolated sample gets sequenced. Pyrosequencing also produces lots and lots of sequencing data very quickly. (One major downside is that each sequence is much shorter than what you get using traditional Sanger Sequencing, by a factor of ten at least. But in this case, the Neanderthal DNA fragments are so short this doesn't matter.)
You can read a really nice explanation of how pyrosequencing works at 454's web site. (For more technical coverage, look here.) Without this technology, a Neanderthal genome project would not be feasible. With this technology, we can now consider all sorts of sequencing projects that would not have been financially or technically feasible before - not just Neanderthal sequencing, but also large scale studies of gene varation in natural populations, including humans. Such large-scale sequencing could help up close in on the genes involved in complex diseases.
I said I was going to talk about sequencing, but I can't resist making a plug for completing the Neanderthal genome. It's interesting to learn about the changes in the genome that took place during evolution, but it's also extremely useful to have a more closely related genome as we try to find and understand the functional portions of the human genome. Having multiple genomes of close species has helped enormously in flies, worms, and yeast (for examples, check out this, this, and this). As is usual in almost any genome-level study of human biology, you can't get far without using an understanding of evolution.
Each group used a different sequencing technology, and as a result, their coverage of the genome differed widely: the Nature group got about 1 million base pairs of sequence, while the Science group got about 65,000 base pairs. (Recall that the human genome contains about 3 billion base pairs, and the Neanderthal genome was undoubtedly similar.) The Neanderthal genome is a challenge because, obviously, any DNA remaining in the 30,000-40,000 year old bones we have is highly fragmented, and the amount of contaminating DNA from microbes (not to mention scientists) is significant. To eventually cover the entire genome, we need a method that can generate lots and lots of sequence without being prohibitively expensive.
The Science group used a more traditional method, which works well for most large-scale genome sequencing efforts, but is not really well-suited for getting huge chunks of Neanderthal genome. As a result, this group obtained only 65,000 base pairs of DNA sequence (which is still a significant accomplishment.) The big problem with this method is that the fragments of DNA isolated from Neanderthal bones have to be cloned before they can be sequenced. (In layman's terms - the DNA fragments have to be placed inside circular pieces of DNA called plasmids, which can be then grown in large quantities inside bactieria.) Many fragments of Neanderthal DNA fail to be cloned at this point, meaning that you lose much of the sample that was painstakingly isolated from the ancient bones.
The Nature group used something called pyrosequencing, which is done on machines called 454 sequencers. Crucially, this technique does not require a cloning step, which means much more of the isolated sample gets sequenced. Pyrosequencing also produces lots and lots of sequencing data very quickly. (One major downside is that each sequence is much shorter than what you get using traditional Sanger Sequencing, by a factor of ten at least. But in this case, the Neanderthal DNA fragments are so short this doesn't matter.)
You can read a really nice explanation of how pyrosequencing works at 454's web site. (For more technical coverage, look here.) Without this technology, a Neanderthal genome project would not be feasible. With this technology, we can now consider all sorts of sequencing projects that would not have been financially or technically feasible before - not just Neanderthal sequencing, but also large scale studies of gene varation in natural populations, including humans. Such large-scale sequencing could help up close in on the genes involved in complex diseases.
I said I was going to talk about sequencing, but I can't resist making a plug for completing the Neanderthal genome. It's interesting to learn about the changes in the genome that took place during evolution, but it's also extremely useful to have a more closely related genome as we try to find and understand the functional portions of the human genome. Having multiple genomes of close species has helped enormously in flies, worms, and yeast (for examples, check out this, this, and this). As is usual in almost any genome-level study of human biology, you can't get far without using an understanding of evolution.
Tuesday, November 21, 2006
Where did all these scientists come from?
It's no secret that it's now more difficult than ever to get funded by the NIH, in spite of the fact that Congress doubled the NIH budget between 1998 and 2003. Researchers have been frustrated and wondering, over coffee and in print, where all the money has gone.
NIH director Elias Zerhouni explains what's going on in the November 17th issue of Science. Grant applications have nearly doubled since 1998 - from 24,151 in 1998 to an expected 46,000 applications in 2006. And this isn't just because individual scientists are applying for more grants - in 1998, 19,000 scientists applied for grants, while in 2006, there were 34,000 scientists who applied.
Where did all these people come from? (I should note that I'm one of these people - I just submitted my first NIH application this summer.) When Congress announced its intention to double the NIH budget, universities started expanding - adding new graduate programs, hiring new faculty, and building new core facilities. All this happened amazingly fast, and now we're feeling the crunch. It doesn't help that the NIH budget hasn't kept pace with inflation since 2003, but Zerhouni presents the numbers that lay to rest other explanations for the funding crunch that have been tossed around - such as an excessive investment in large clinical trials or big, Manhattan project-style science at the expense of smaller, innovative projects initiated by individual researchers.
Is this a good thing? The down side is that with more people we'll get more fraud, more mediocre science, and more fragmentation of the scientific community. It's already barely possible to seriously keep up with the literature in one's own field - which means it will be harder to find people on review committees who understand each other. It's much, much harder for a young investigator to get started - in the past, scientists in their 20's and early 30's have been among the most innovative and creative scientists, but young scientists today have their motivation and creativity squashed by the high barriers to independence, barriers which are only overcome when some of your best years are over with. Sure, older scientists are still damn good researchers, but if the start of your scientific career is creatively stunted, it can hobble your thinking later on.
In spite of these drawbacks, the fact is that there is still a hell of a lot of good scientific work to do, even if all of it isn't the most pathbreaking or innovative science. There are a lot of useful details to be worked out, enough to keep people busy for a long time. Money invested in new scientists will be money well spent - much more well spent than much of billions of dollars we lost in the attempted reconstruction of Iraq, money which did more to enrich the already bloated pockets of Dick Cheney's friends than it did to benefit the Iraqis.
The investment in research infrastructure made by US research universities and biotech companies in recent years has helped keep the US at the forefront of an increasingly competitive world-wide scientific community. If we want to stay there, we need to pay for it.
As Zerhouni put it:
"Since 1945, United States success in scientific research and development has been the result of the implicit partnership that exists among academia, the federal government, and industry. In this model, research institutions take the risk of building and developing our national scientific capacity; the federal government, through a competitive peer-review process, funds the best science; and industry plays the critical role of bringing new, safe, and effective products to the public. This strategy is the keystone to sustaining American competitiveness, and must be preserved."
NIH director Elias Zerhouni explains what's going on in the November 17th issue of Science. Grant applications have nearly doubled since 1998 - from 24,151 in 1998 to an expected 46,000 applications in 2006. And this isn't just because individual scientists are applying for more grants - in 1998, 19,000 scientists applied for grants, while in 2006, there were 34,000 scientists who applied.
Where did all these people come from? (I should note that I'm one of these people - I just submitted my first NIH application this summer.) When Congress announced its intention to double the NIH budget, universities started expanding - adding new graduate programs, hiring new faculty, and building new core facilities. All this happened amazingly fast, and now we're feeling the crunch. It doesn't help that the NIH budget hasn't kept pace with inflation since 2003, but Zerhouni presents the numbers that lay to rest other explanations for the funding crunch that have been tossed around - such as an excessive investment in large clinical trials or big, Manhattan project-style science at the expense of smaller, innovative projects initiated by individual researchers.
Is this a good thing? The down side is that with more people we'll get more fraud, more mediocre science, and more fragmentation of the scientific community. It's already barely possible to seriously keep up with the literature in one's own field - which means it will be harder to find people on review committees who understand each other. It's much, much harder for a young investigator to get started - in the past, scientists in their 20's and early 30's have been among the most innovative and creative scientists, but young scientists today have their motivation and creativity squashed by the high barriers to independence, barriers which are only overcome when some of your best years are over with. Sure, older scientists are still damn good researchers, but if the start of your scientific career is creatively stunted, it can hobble your thinking later on.
In spite of these drawbacks, the fact is that there is still a hell of a lot of good scientific work to do, even if all of it isn't the most pathbreaking or innovative science. There are a lot of useful details to be worked out, enough to keep people busy for a long time. Money invested in new scientists will be money well spent - much more well spent than much of billions of dollars we lost in the attempted reconstruction of Iraq, money which did more to enrich the already bloated pockets of Dick Cheney's friends than it did to benefit the Iraqis.
The investment in research infrastructure made by US research universities and biotech companies in recent years has helped keep the US at the forefront of an increasingly competitive world-wide scientific community. If we want to stay there, we need to pay for it.
As Zerhouni put it:
"Since 1945, United States success in scientific research and development has been the result of the implicit partnership that exists among academia, the federal government, and industry. In this model, research institutions take the risk of building and developing our national scientific capacity; the federal government, through a competitive peer-review process, funds the best science; and industry plays the critical role of bringing new, safe, and effective products to the public. This strategy is the keystone to sustaining American competitiveness, and must be preserved."
Wednesday, November 15, 2006
How should we teach our kids math?
The NY Times is reporting on the lagging math skills of US kids and efforts to change, yet again, how we teach math. I'm sympathetic to the desire to teach math in a way that doesn't turn people off. Too many people (including some people who grow up to be biologists) go through their education feeling very, very insecure about their ability to do and understand math.
What's tough about this problem is that you just can't teach the concepts and let kids figure out for themselves how to solve the problems. Understanding is good, but in math, understanding is not a substitute for the ability that comes with lots and lots of practice. This is different from many other fields of study, where if you understand the basic ideas and arguments you can work out a lot for yourself. To actually do math well, you need regular, sometimes mind-numbing practice - you can't just reinvent the wheel (i.e., derive your results from first principles) every time you need to solve a problem.
Gaining proficiency in math is similar to being able to do the NY Times crossword puzzle or play arpeggios on the piano - you simply need a lot of repetitive practice. You may know what an arpeggio is, but that's different from being able to play them over the entire keyboard, in all major and minor keys at a fast tempo.
Just like with the piano, kids will like math more when they are actually reasonably good at it. And there is no reason that most kids can't be fairly good at the kind of basic math we'd expect every educated person to know. Our teaching needs to reflect that - it's good to encourage understanding, but proficiency will never come without plenty of practice.
What's tough about this problem is that you just can't teach the concepts and let kids figure out for themselves how to solve the problems. Understanding is good, but in math, understanding is not a substitute for the ability that comes with lots and lots of practice. This is different from many other fields of study, where if you understand the basic ideas and arguments you can work out a lot for yourself. To actually do math well, you need regular, sometimes mind-numbing practice - you can't just reinvent the wheel (i.e., derive your results from first principles) every time you need to solve a problem.
Gaining proficiency in math is similar to being able to do the NY Times crossword puzzle or play arpeggios on the piano - you simply need a lot of repetitive practice. You may know what an arpeggio is, but that's different from being able to play them over the entire keyboard, in all major and minor keys at a fast tempo.
Just like with the piano, kids will like math more when they are actually reasonably good at it. And there is no reason that most kids can't be fairly good at the kind of basic math we'd expect every educated person to know. Our teaching needs to reflect that - it's good to encourage understanding, but proficiency will never come without plenty of practice.
Sunday, November 12, 2006
The problem with computational biology papers
OK, my title is too general - it should be, "The problem with some computational biology papers that deal with certain research questions." There is a type of trendy science that frequently crops up in many journals (including good ones like Nature and Science. It basically goes like this (for a prime example, look here):
1. A computational biology lab sees one or more genomic-scale datasets that they can do some calculations on (usually microarray data).
2. The computational biologists come up with some algorithm that's supposedly better than what's out there, and they crunch the numbers on the genomic data. This results in some predictions of novel regulatory interactions - for example, they predict that certain transcription factors regulate certain genes involved in cell division. At this point we have no idea whether their predictions are right, or even persusive enough to be worth testing. But it's a start.
3. The computationl biologists "validate" their results by using (notoriously incomplete) database annotations about the genes in their predictions, or by a shallow, cursory scan of the experimental literature (which the authors are usually not that familiar with). They then state something like "75% of our predicted transcription factor-gene interactions have some basis in the literature." Up to this point things are fine (they have made predictions, and given us some reason to believe that the predictions have a chance of being right), but then they usually go on and say something like this: "Therefore, we have demonstrated that our algorithm has the ability to find new regulatory interactions..." They have demonstrated no such thing. They have made predictions, but haven't bothered to test them; instead, they do a crappy literature survey (usually with significant omissions).
The result is that you get different groups coming up with all sorts of new analyses of the same genomic data (in my field, cell cycle gene expression and genome-wide transcription factor binding data are big ones), but never really making any serious progress towards improving our understanding of the biological process in question. The worst part is that, over time, the researchers doing this kind of work start talking as if we are making progress in our understanding, even though we haven't really tested that understanding. You start getting an echo chamber resonating with these guys who are citing each other for validation more than they are citing the people actually study the relevant genes in the lab.
This means that the experimentalists ignore the echo chamber, and then computational biology becomes irrelevant to experimental biology - which is a sad thing. There are so many 'validated' predictions out there, that the experimentalists don't really know where to start, where the good predictions are. And the computational researchers don't care enough to really work with someone who will actually go test things in the lab, in spite of the fact that if these computational biologists did care enough, they would get more notice from the experimentalists.
The problem is bad enough that one journal, Nucleic Acids Research changed their policy on computational papers:
"Computational biology
Manuscripts will be considered only if they describe new algorithms that are a substantial improvement over current applications and have direct biological relevance. The performance of such algorithms must be compared with current methods and, unless special circumstances prevail, predictions must be experimentally verified. The sensitivity and selectivity of predictions must be indicated. Small improvements or modifications of existing algorithms will not be considered. Manuscripts must be written so as to be understandable to biologists. The extensive use of equations should be avoided in the main text and any heavy mathematics should be presented as supplementary material. All source code must be freely available upon request."
This is a move in the right direction. But until more journals adopt this stance, beware of researchers who claim to have calculated the gene regulatory network for this or that process, or have identified 'modules' of interacting proteins that perform a function in the cell. If these claims, usually based on noisy, less than ideal genomic data, haven't been tested with serious experiments, they remain unproven hypotheses.
1. A computational biology lab sees one or more genomic-scale datasets that they can do some calculations on (usually microarray data).
2. The computational biologists come up with some algorithm that's supposedly better than what's out there, and they crunch the numbers on the genomic data. This results in some predictions of novel regulatory interactions - for example, they predict that certain transcription factors regulate certain genes involved in cell division. At this point we have no idea whether their predictions are right, or even persusive enough to be worth testing. But it's a start.
3. The computationl biologists "validate" their results by using (notoriously incomplete) database annotations about the genes in their predictions, or by a shallow, cursory scan of the experimental literature (which the authors are usually not that familiar with). They then state something like "75% of our predicted transcription factor-gene interactions have some basis in the literature." Up to this point things are fine (they have made predictions, and given us some reason to believe that the predictions have a chance of being right), but then they usually go on and say something like this: "Therefore, we have demonstrated that our algorithm has the ability to find new regulatory interactions..." They have demonstrated no such thing. They have made predictions, but haven't bothered to test them; instead, they do a crappy literature survey (usually with significant omissions).
The result is that you get different groups coming up with all sorts of new analyses of the same genomic data (in my field, cell cycle gene expression and genome-wide transcription factor binding data are big ones), but never really making any serious progress towards improving our understanding of the biological process in question. The worst part is that, over time, the researchers doing this kind of work start talking as if we are making progress in our understanding, even though we haven't really tested that understanding. You start getting an echo chamber resonating with these guys who are citing each other for validation more than they are citing the people actually study the relevant genes in the lab.
This means that the experimentalists ignore the echo chamber, and then computational biology becomes irrelevant to experimental biology - which is a sad thing. There are so many 'validated' predictions out there, that the experimentalists don't really know where to start, where the good predictions are. And the computational researchers don't care enough to really work with someone who will actually go test things in the lab, in spite of the fact that if these computational biologists did care enough, they would get more notice from the experimentalists.
The problem is bad enough that one journal, Nucleic Acids Research changed their policy on computational papers:
"Computational biology
Manuscripts will be considered only if they describe new algorithms that are a substantial improvement over current applications and have direct biological relevance. The performance of such algorithms must be compared with current methods and, unless special circumstances prevail, predictions must be experimentally verified. The sensitivity and selectivity of predictions must be indicated. Small improvements or modifications of existing algorithms will not be considered. Manuscripts must be written so as to be understandable to biologists. The extensive use of equations should be avoided in the main text and any heavy mathematics should be presented as supplementary material. All source code must be freely available upon request."
This is a move in the right direction. But until more journals adopt this stance, beware of researchers who claim to have calculated the gene regulatory network for this or that process, or have identified 'modules' of interacting proteins that perform a function in the cell. If these claims, usually based on noisy, less than ideal genomic data, haven't been tested with serious experiments, they remain unproven hypotheses.
Wednesday, November 01, 2006
A Republican War on Science? Nature's editors cop out
The Oct 19th issue of Nature contains a feature section on science and the upcoming US Conrgessional elections. In one of the editorials, (subscription required) Nature's editors criticize the phrase "Republican war on science":
"Slogans such as the 'Republican war on science', meant to sum up a host of perceived abuses, do not do justice to the complex relationship between science and each of the two major political parties."
In an effort to not be percieved as partisan, some people, Nature's editors included, can't bring themselves to truly call things as they are. In recent years, there are very good reasons to single out the Republican party for its serious corrosion of the US government's relationship with science. The phrase "Republican War on Science" (the title of Chris Mooney's recent book) is a correct, legitimate characterization for the following reasons:
1. It's true that no political party or presidential administration is monolithic. There are many Republicans who are not part of an assault on the integrity of science, and not every single decision made by the Bush adminstration has been bad for science. However, the Republican leadership in Congress and the Executive Branch, as well as the active members of the Republican base, have seriously abused science and scientists to push their ideological agenda. Whether it's pushing intelligent design (from school boards to the 'Santorum Amendment' of the No Child Left Behind Act), having an ex-physician novelist testify to Congress on climate change, diagnosing Terri Schiavo by video from Congress, or the more low-profile but perasive agency decisions that weaken protection of the environment and endangered species, promote ineffective and sometimes inaccurate 'abstinence only' sex ed programs, and restrict drugs because of anti-abortion ideology and not safety and efficacy concerns, the Republican leadership and base have attacked mainstream, scientific consensus when it stands in the way of their ideological postion. In recent years, Republicans have been much, much more guilty of this than Democrats.
2. Yes, the Republican-led Congress voted to double the NIH budget in the late 90's, and recently voted to double the NSF budget. But this is an easy vote - it doesn't offend anyone's ideology, it's a fairly small fraction of overall governtment spending on R&D, and there is large bipartisan support for these increases. These votes don't negate the Congressional meddling in research whenever that research is politically controversial for conservatives.
3. This 'war on science' fits the description given by Rep. Rush Holt of the current political climate (in Nature, subscription required):
"In official Washington, scientific subjects have become really politicized. There should be debate about the policy that is derived from science. But, historically, if science puts limits on the choices that are possible, the politicians would accept that. Now, by treating science as just another topic to be dealt with ideologically, or to be part of political trades, they will even ignore the laws of science."
Nature's editors should have shown some spine on this issue. At the very least, they shouldn't have taken a blatant swipe at Chris Mooney, who has been a serious champion of scientific integrity in both journalism and government.
"Slogans such as the 'Republican war on science', meant to sum up a host of perceived abuses, do not do justice to the complex relationship between science and each of the two major political parties."
In an effort to not be percieved as partisan, some people, Nature's editors included, can't bring themselves to truly call things as they are. In recent years, there are very good reasons to single out the Republican party for its serious corrosion of the US government's relationship with science. The phrase "Republican War on Science" (the title of Chris Mooney's recent book) is a correct, legitimate characterization for the following reasons:
1. It's true that no political party or presidential administration is monolithic. There are many Republicans who are not part of an assault on the integrity of science, and not every single decision made by the Bush adminstration has been bad for science. However, the Republican leadership in Congress and the Executive Branch, as well as the active members of the Republican base, have seriously abused science and scientists to push their ideological agenda. Whether it's pushing intelligent design (from school boards to the 'Santorum Amendment' of the No Child Left Behind Act), having an ex-physician novelist testify to Congress on climate change, diagnosing Terri Schiavo by video from Congress, or the more low-profile but perasive agency decisions that weaken protection of the environment and endangered species, promote ineffective and sometimes inaccurate 'abstinence only' sex ed programs, and restrict drugs because of anti-abortion ideology and not safety and efficacy concerns, the Republican leadership and base have attacked mainstream, scientific consensus when it stands in the way of their ideological postion. In recent years, Republicans have been much, much more guilty of this than Democrats.
2. Yes, the Republican-led Congress voted to double the NIH budget in the late 90's, and recently voted to double the NSF budget. But this is an easy vote - it doesn't offend anyone's ideology, it's a fairly small fraction of overall governtment spending on R&D, and there is large bipartisan support for these increases. These votes don't negate the Congressional meddling in research whenever that research is politically controversial for conservatives.
3. This 'war on science' fits the description given by Rep. Rush Holt of the current political climate (in Nature, subscription required):
"In official Washington, scientific subjects have become really politicized. There should be debate about the policy that is derived from science. But, historically, if science puts limits on the choices that are possible, the politicians would accept that. Now, by treating science as just another topic to be dealt with ideologically, or to be part of political trades, they will even ignore the laws of science."
Nature's editors should have shown some spine on this issue. At the very least, they shouldn't have taken a blatant swipe at Chris Mooney, who has been a serious champion of scientific integrity in both journalism and government.
Saturday, October 28, 2006
Classic papers in quantitative biology
The hot trend in biology education right now is to train undergrads, grad students, and postdocs in not only biology, but also in enough math, computer science, and physics so that they can function successfully in the highly interdisciplinary research fields that are starting play a more important role in current research. Most major research universities are spending serious time and money thinking about how to help physicists and computer scientists become competent in biology, and biologists become competent in the relevant physics and computer science. We have a program for this purpose here at Washington University.
Biologists at Princeton have been experimenting with a variety of educational approaches, and in this month's issue of Nature Reviews Molecular Cell Biology has an essay about one course at Princeton that focuses on what I'd call 'classic papers in quantitative biolgy.' They put their grad students in a room - physicists, biologists, etc., all interested in biological problems, and have them discuss papers that contain not only great biology, but great examples of quantitative work as well.
Anyway, in this essay, there is a list of 12 of the best papers used in the course. If you're interested at all in genomics or what's being called systems biology, I highly recommend checking out this essay and the list of papers. Unfortunately you need some sort of university library access for most of them, but they're worth the effort to get them.
Biologists at Princeton have been experimenting with a variety of educational approaches, and in this month's issue of Nature Reviews Molecular Cell Biology has an essay about one course at Princeton that focuses on what I'd call 'classic papers in quantitative biolgy.' They put their grad students in a room - physicists, biologists, etc., all interested in biological problems, and have them discuss papers that contain not only great biology, but great examples of quantitative work as well.
Anyway, in this essay, there is a list of 12 of the best papers used in the course. If you're interested at all in genomics or what's being called systems biology, I highly recommend checking out this essay and the list of papers. Unfortunately you need some sort of university library access for most of them, but they're worth the effort to get them.
Thursday, October 26, 2006
Comparing Genomes on a Chip
Last week at our lab meeting I had a chance to hear a presentation by Doug Berg, a microbiologist here at Washington University. Berg's work is a great combination of new technology, genomics and evolution, and it happens to also have potential medical relevance. He's studying the evolution of drug resistance in Helicobacter pylori, a ususally benign bacterium that is responsible for stomach ulcers. (Recall that the Nobel Prize in medicine was awarded last year to Barry Marshall and Robin Warren for their discovery of the link between H. pylori and ulcers.)
Berg is basically evolving highly drug resistant bacteria in the lab and using a new genome comparison technology to identify the genes that are changed as the bacterial strains become more resistant. The drug used in these experiments is metronidazole - a substance that itself is not harmful, but inside a bacterial cell it is metabolized into a toxin that causes significant DNA damage.
To create these drug-resistant strains, bacteria are spread on an agar plate - a peteri dish filled with a Jello-like substance that contains nutrients and metronidazole. Most bacteria are killed by the drug, but some individual bacteria survive, if they harbor random mutations that just happen to confer drug resistance. These surviving bacteria continue to divide, and after a few days a colony - a blob of bacteria all descended from the original surviving bacterium - has formed on the plate. You can pick off this colony with a toothpick and save it; you now have a drug resistant strain of bacteria. This process can be repeated - grow up the drug resistant strain, smear it on an agar plate with a higher concentration of the drug, and watch for the next round of colonies to grow from the survivors.
After doing many experiments like this, the Berg lab gets a series of H. pylori strains, each with varying drug resistance. The obvious question then is, how do the genomes of these strains differ? What specific (but randomly occuring) mutations had to take place in order for this level of drug resistance to evolve? Sequencing the genomes of each of these various drug-resistant strains would be the obvious way to find these differences, but doing that kind of sequencing is still prohibitively expensive and computationally intentive. But the Berg lab is using a new technique that rapidly compares genomes on a chip. (The link leads to an abstract of their paper; the full text is subscription only but you can check out more about the technique in a "webinar" found here, at the bottom of the page.). They use microarrays (DNA chips) that cover the entire genome (called a 'tiling array'). By hybridizing DNA from both the nomral and drug resistant strains to the microarray (check out my earlier summary of how a microarray works), they can identify those spots on the array (and thus in the genome) where the two strains differ.
Using this technique, the Berg lab has identified key genes involved in H. pylori resistance to metronidozole. There seem to be some genes that are always changed in these drug resistant strains, and some that can be different. In other words, there seem to be multiple evolutionary paths to drug resistance, but those paths often cross.
This techique of comparing genomes on a chip has a lot of potential, and within a few years, something like this may start showing up in common medical care and hospital labs, replacing the more primitive techniques that are currently used for classifying tumors or drug-resistant pathogens.
Berg is basically evolving highly drug resistant bacteria in the lab and using a new genome comparison technology to identify the genes that are changed as the bacterial strains become more resistant. The drug used in these experiments is metronidazole - a substance that itself is not harmful, but inside a bacterial cell it is metabolized into a toxin that causes significant DNA damage.
To create these drug-resistant strains, bacteria are spread on an agar plate - a peteri dish filled with a Jello-like substance that contains nutrients and metronidazole. Most bacteria are killed by the drug, but some individual bacteria survive, if they harbor random mutations that just happen to confer drug resistance. These surviving bacteria continue to divide, and after a few days a colony - a blob of bacteria all descended from the original surviving bacterium - has formed on the plate. You can pick off this colony with a toothpick and save it; you now have a drug resistant strain of bacteria. This process can be repeated - grow up the drug resistant strain, smear it on an agar plate with a higher concentration of the drug, and watch for the next round of colonies to grow from the survivors.
After doing many experiments like this, the Berg lab gets a series of H. pylori strains, each with varying drug resistance. The obvious question then is, how do the genomes of these strains differ? What specific (but randomly occuring) mutations had to take place in order for this level of drug resistance to evolve? Sequencing the genomes of each of these various drug-resistant strains would be the obvious way to find these differences, but doing that kind of sequencing is still prohibitively expensive and computationally intentive. But the Berg lab is using a new technique that rapidly compares genomes on a chip. (The link leads to an abstract of their paper; the full text is subscription only but you can check out more about the technique in a "webinar" found here, at the bottom of the page.). They use microarrays (DNA chips) that cover the entire genome (called a 'tiling array'). By hybridizing DNA from both the nomral and drug resistant strains to the microarray (check out my earlier summary of how a microarray works), they can identify those spots on the array (and thus in the genome) where the two strains differ.
Using this technique, the Berg lab has identified key genes involved in H. pylori resistance to metronidozole. There seem to be some genes that are always changed in these drug resistant strains, and some that can be different. In other words, there seem to be multiple evolutionary paths to drug resistance, but those paths often cross.
This techique of comparing genomes on a chip has a lot of potential, and within a few years, something like this may start showing up in common medical care and hospital labs, replacing the more primitive techniques that are currently used for classifying tumors or drug-resistant pathogens.
NY Times feature on science fraud
The NY Times has a feature article about the first case of scientific fraud with NIH money that resulted in a jail sentence for the scientist involved. A young technician in the lab of Eric Poehlman risked his reputation and future career to expose years of fraudulent research in the Poehlman lab. The grad students and postdocs in the lab were willing to look the other way for years, even though they knew the data that was ending up in the lab's published paper's sometimes came straight out of Poehlman's imagination.
I'm glad Poehlman went to prison. Someone who commits massive fraud like that wrecks trust within the scientific community and with the greater public. Without trust, between colleagues, and between the public and the scientists paid by the public, the whole merit-based funding system is threatened.
I'm glad Poehlman went to prison. Someone who commits massive fraud like that wrecks trust within the scientific community and with the greater public. Without trust, between colleagues, and between the public and the scientists paid by the public, the whole merit-based funding system is threatened.
Friday, October 20, 2006
Can you learn lab skills with a keyboard and mouse, instead of pipettes and test tubes?
Today's NY Times features an article about a conflict among educators over online lab courses. Universities are debating whether to accept virtual lab courses as a substitute for real Freshman science labs, especially when it comes to AP credit.
Online schools allow students to do virtual pig dissections, virtual DNA gels, and virtual chemistry experiments, and defenders of the vitrual lab courses argue that their students do just as well, if not better on AP exams. If your goal is a high exam score, then I don't question that these virtual labs can help students effectively learn the concepts. But science is more than just concepts, and virtual labs can never take the place of real labs in seriously training scientists.
Learning to be a scientist is much like an apprenticeship - you could never learn to be a master chef, violin maker, or serious gardner if you tried to learn all your skills in an online simulation, even though that simulation might help you learn the concepts involved. You can't learn to be a serious mechanical engineer if you never set foot in a machine shop. Experimental science requires something like a green thumb - a hard to desciribe ability to use your tools with just the right touch. Many experiments are technically challenging, and require dexterity and experience to do them well - that is, reproducibly. (And, going back to the NY Times article, the 'kitchen chemistry' they describe is no substitute. Kitchen chemistry is usually just a bunch of magic tricks; it doesn't come close to teaching kids what real experiments are like.)
Actually, I think this point is generally true of all online learning that tries to be a substitute for a real univerity education. All serious scholarship, whether it involves lab work or not, requires an element of apprenticeship to learn. At most, virtual courses can help students learn the basics, but a complete online university is more like a technical school or certification program, not a real university.
But what about kids in poor schools that can't afford to have serious science labs for their AP courses? In those cases, a good virtual lab is probably better than a poor or nonexistent real lab. Universities have to decide, whether all of their graduates need that kind of lab experience. For students graduating in science or medical fields, the answer is an unequivocal yes. For the others, I don't know the answer. But, in light of the fact that the US is producing fewer and fewer home-grown scientists, a great hands-on science lab can be a great way to get the AP students who took these virtual lab classes interested in a science career.
Online schools allow students to do virtual pig dissections, virtual DNA gels, and virtual chemistry experiments, and defenders of the vitrual lab courses argue that their students do just as well, if not better on AP exams. If your goal is a high exam score, then I don't question that these virtual labs can help students effectively learn the concepts. But science is more than just concepts, and virtual labs can never take the place of real labs in seriously training scientists.
Learning to be a scientist is much like an apprenticeship - you could never learn to be a master chef, violin maker, or serious gardner if you tried to learn all your skills in an online simulation, even though that simulation might help you learn the concepts involved. You can't learn to be a serious mechanical engineer if you never set foot in a machine shop. Experimental science requires something like a green thumb - a hard to desciribe ability to use your tools with just the right touch. Many experiments are technically challenging, and require dexterity and experience to do them well - that is, reproducibly. (And, going back to the NY Times article, the 'kitchen chemistry' they describe is no substitute. Kitchen chemistry is usually just a bunch of magic tricks; it doesn't come close to teaching kids what real experiments are like.)
Actually, I think this point is generally true of all online learning that tries to be a substitute for a real univerity education. All serious scholarship, whether it involves lab work or not, requires an element of apprenticeship to learn. At most, virtual courses can help students learn the basics, but a complete online university is more like a technical school or certification program, not a real university.
But what about kids in poor schools that can't afford to have serious science labs for their AP courses? In those cases, a good virtual lab is probably better than a poor or nonexistent real lab. Universities have to decide, whether all of their graduates need that kind of lab experience. For students graduating in science or medical fields, the answer is an unequivocal yes. For the others, I don't know the answer. But, in light of the fact that the US is producing fewer and fewer home-grown scientists, a great hands-on science lab can be a great way to get the AP students who took these virtual lab classes interested in a science career.
Sunday, October 15, 2006
Reasons to oppose stem cell research? Debunking the opposition to Missouri's Amendment 2
Around my neighborhood here in St. Louis you can find a lot of lawn signs dealing with "Amendment 2" - a stem cell research and therapies amendment which will be on the ballot next month.
As a general principle, I think the public (the taxpayers funding scientific research in the US) should be involved discussing what kind of science should be done in our society. Mind you, they shouldn't be judging the scientific, technical merit of specific proposals - that's as absurd as asking someone with no engineering training to evaluate the structural integrity of a proposed bridge design.
But some general input is good. I just submitted a funding proposal to the American Cancer Society. On their review panels, they include non-scientists who are interested in cancer research, and my application includes an explanation in non-technical language of what I'm proposing to do and how it is related to cancer. I am judged partly on how well I can explain my work to a general audience, and, more importantly, on whether I can persuade the non-scientist on the panel that my work is important for cancer research. I think that's a great part of the process.
On the other hand, often in the political arena, people's arguments on science issues are based on pure dishonesty and bad faith. The opposition movement to Missouri's amendment 2 falls under this category; they are lying to scare people into voting against the amendment. In their flyers and on their website, they lecture us about ethics, but they are liars and deserve no credibility on this issue.
These are harsh words, but as you'll see below, almost every reason they give to vote against this bill is either deliberately misleading or based on a flat-out lie.
To see that these people are liars, you have to know first what the amendment says. It's never a good idea to take someone else's word on what a ballot initiative says, so you read it yourself right here. And if you're not a resident of Missouri, you should still be interested, because many other states are wrestling with the same issue right now, not to mention our national debate on the subject.
Here's my summary of what I think is a very reasonable initiative (and really, check me by reading the initiative yourself):
The Main Point: stem cell research and therapies permitted under federal law shall be legal in Missouri. Since very little is actually legal or funded under current federal law (and this amendment isn't providing any money), right now this makes little difference. This amendment is really aimed at a time when, say, Congress could override a Bush veto of a stem cell bill like the one that was recently vetoed. This amendment is just assuring that stem cell research and therapies are treated like any other research. You don't generally see certain mainstream research or therapies banned in certain states, while permitted overall at the federal level.
Important restrictions specified in the amendment:
- no cloning a human being - that is, you can't implant a human embryo in a human uterus, if that embryo was not created with human sperm and egg. A cloned embryo is created without sperm (referred to below as Somatic Cell Nuclear Transfer or SCNT).
- no creating a blastocyst purely for research by fertilization - in other words, embryos taken for stem cells would be left over at in vitro fertilization (IVF) clinics, or created through producing a clone by SCNT (if permitted by federal law).
-no taking cells from a blastocyst after more than 14 days of cell division
-no selling eggs or blastocysts
The amendment also includes typical provisions that already apply to all ongoing research using human subjects or samples; these provisions are redundant, because they are already required by current federal laws and regulations:
- donors of eggs/blastocysts must give informed, voluntary consent
- researchers must comply with normal standards of bioethics, must have Institutional Review Board approval (required at all research universities before someone can work with human subjects or tissue samples), and comply with all other regulations that apply to research with human subjects and samples in general.
This proposal basically deals with all non-fundamentalist ethical issues, and I predict that such an approach will be adopted at the federal level within 4 years (and maybe even during the next Congressional session).
A side note on cloning: Beware of people who try to tell you cloning is cloning is cloning - and that there is no difference, it's all ethically the same. Not true! Scientists use the term cloning to mean different things. Cloning a gene is not the same a cloning a human being. Molecular biologists make copies of genes and put them into a variety of different contexts; they call that cloning. Making a human clone involves implanting a cloned embryo into a uterus, and having the woman eventually deliver the baby. This has not been done yet, and it's unethical because you would probably have hundreds of women suffer hundreds of miscarriages before you actually had a live birth. On top of that, that live child will probably have future health problems (because that's what we've observed in cloned animals). But these human clones would not be soulless robots (the Hollywood-inspired fear expressed by a woman I met on the train the other month) - they would be like the natural clones among us today - identical twins, fully autonomous human beings.
Cloning embryos for stem cell research starts out the same as human cloning (pull the DNA from an egg, put in DNA from an adult cell, and get it to divide without fertilization by sperm - SCNT), but you don't implant the embryo in a uterus. People worry that doing this though is the first step towards human cloning, and what's to stop us from going all the way? Well, the fact that cloning an embryo is the easy part, getting it to develop in the uterus is the really hard part. And also, the ethical issues significantly change once you implant such an embryo in the uterus, as I stated before.
Back to amendment 2: the proposed amendment is facing a disinformation campaign by a dishonest opposition. The "Missourians Against Human Cloning" have put out 20 talking points, each of which is either severely misleading or an outright lie. If you've had enough of this issue, you can stop reading the post here, but for all the gory details, keep reading:
- "Reason 1: Amendment 2 would permanently change Missouri's Constitution." Permanently, until voters adopt an amendment to change this one. This claim is misleading, because it ignores that fact that, unlike what happens at the federal level, state constitutional amendments are a common way of doing business in most states, and the amendments are easy to change. Missouri has four proposed amendments on the ballot this Fall, most dealing with mundane issues like property tax exemptions and pensions for state officials convicted of felonies. There is nothing unusual about putting an issue like stem cell research into a state constitutional amendment.
- "Reason 2: Amendment 2 will use our tax dollars for unethical and unproven research. Despite claims to the contrary, if Amendment 2 passes, Missourians will pay for unethical and unproven research indefinitely." Also dishonest - the amendment provides NO MONEY. Any money for such research would come from the NIH budget, which is unlikely to get an extra raise specifically for stem cell research. If you want to talk about our tax dollars going to something unethical (and, of unproven or disproven effectiveness), let's talk about the Bush administration's torture policy.
- "Reason 3: Amendment 2 would create a constitutional right to devalue human life - it would treat human life as a commodity and raw material for unethical human experimentation by the bio-tech industry." Here's an outright lie - the amendment specifically prohibits treating eggs or embryos as commodities - that is, something to be bought and sold. Beyond that, the claim that stem cell research for disease cures in some vague, unarticulated way devalues human life is weak. Notice the scare language that implicitly evokes Nazi-style research - stem cell research is 'unethical human experimentation'!
- "Reason 4 - Amendment 2 is harmful to women... up to 35% of women who submit to ovarian stimulation experience health consequences, and up to 14% of these are severe." Those numbers are false. The procedure is uncomfortable, but it's not really more risky than an appendectomy. It's done routinely, every day, all over this country. My wife went through many rounds of ovarian stimulation drugs, and went through egg retrieval twice. These drugs have been used for a long time, they have a long track record, and they are safe in the vast majority of cases (no riskier than the anesthesia used in a routine appendectomy). It's extremely common, and I would be money that you know someone who has taken ovarian stimulation drugs, even though you may not be aware of it.
- "Reason 5 - Amendment 2 is unnecessary - adult stem cell research has been proven effective for many illnesses... We need to retain the option to direct our tax dollars to this ethical and promising research." That's false - there are actually few illnesses that adult stem cells work for - anything requiring a bone marrow transplant, and that's just about it. We always have the option to study adult stem cells, and labs who are already doing this work are not suddenly going to drop it once there is money for embryonic stem cell research. Furthermore, the adult stem cell researchers I know personally are some of the most passionate advocates for embryonic stem cell research, even though they have no intention of dropping all of their current work to study embryonic stem cells should funding become available.
- "Reason 6: Amendment 2 protects and promotes human cloning. The proponents want you to believe that Somatic Cell Nuclear Transfer (SCNT) is NOT cloning, but the reality is that every textbook and scientific journal defines SCNT as Cloning." Here they dishonestly try to muddy the waters by exploiting the fact that scientists use the term cloning in different ways - see my earlier note on cloning. The ethical issues relevant to making a live human clone are different from those involving cloning embryos (or SCNT) that will never see the inside of a uterus. You have to articulate each of these ethical cases individually. I can think of reasons why it's wrong to make human clones (health problems in the cloned humans, emotional and physical risks to the women who carry these pregnancies with a high risk of failure), but those reasons don't apply to embryos that are never implanted in a uterus. The Amendment 2 opponents are think they can just get away with screaming CLONING!!! instead of articulating a serious argument against embryo cloning. This is a tactic that repeatedly shows up in this debate.
- "Reason 7 Amendment 2 is an elite initiative" - basically, 95% of the promotional money for this amendment came from one wealthy couple. This talking point is not dishonest, it's just stupid. We'll see how elite this amendment really is when we vote on it next month.
- "Reason 8 - Amendment 2 is immoral. It is ethically wrong to destroy human life regardless of its origin or geography." I'm not sure what geography has to do with anything here, but if Amendment 2 is immoral because it 'destroys human life', then so is the process used for in vitro fertilization and many common forms of birth control. Most citizens don't seem to be outraged over birth control or IVF, so I take it that most people don't buy this ethical reasoning.
- "Reason 9 - Amendment 2 will exploit the disadvantaged" because they will be paid to donate their eggs for research. In reality, this won't exploit the 'disadvantaged' any more than any other research involving human subjects - people who volunteer for such studies are paid a small fee in return for their time, effort, and travel expenses. This is how all new drugs and treatments are tested - if amendment 2 is exploitative, so is essentially all research with human subjects. Many such studies involve procedures much more risky than egg donation.
- "Reason 10: Amendment 2 is wrong. To permanently change our constitution to protect unproven and risky scientific experimentation is wrong." We're only halfway to Reason 20, and already they're recycling their previous reasons (see Reasons 1, 2, and 5). I guess this means that there really aren't 20 independent reasons to vote against Amendment 2.
- "Reason 11: Amendment 2 ignores proven research. Ethical stem cell research provides real hope without cloning or destroying human life..." Now they're recycling Reason 5.
- "Reason 12: Amendment 2 is deceptive - it will PROMOTE not ban human cloning." Recycling Reason 6 here. Like I said above, this is a dishonest and misleading attempt to muddy the various meanings of 'cloning'. Amendment 2 bans cloning human beings, but it does not ban the creation of cloned human embryos that will never be implanted in a uterus. Amendment 2 is clear about it's definitions (again, go read the entire thing yourself); the Missourians Against Human Cloning are the ones being deceptive.
- "Reason 13: Amendment 2 will destroy human life." Amendment 2 could save human lives through the treatments it might facilitate. Unless you think destroying blastocysts is equal to murder, Reason 13 is not true.
- "Reason 14: Amendment 2 will NOT give Missourians more access rights to cures or therapies that they and every other American ALREADY enjoy." This is dishonest - that's not the point of the amendment; the point is to make sure that future therapies permitted elsewhere aren't banned in Missouri.
- "Reason 15: Amendment 2 ignores the success of Adult Stem Cell therapies and cures." Wow, we're recycling Reason 5 for a second time. Amendment 2 does not 'ignore' Adult stem cell research - it doesn't ban it and it doesn't take away research dollars (especially since it doesn't provide any research dollars for embryonic stem cell research). And naturally adult stem cells therapies are farther along than embryonic stem cell therapies - adult stem cell research has been going on longer, and that research hasn't been blocked at the federal level.
- "Reason 16: Amendment 2 will NOT insure First Class status to Missouri's biotech industry." This is, amazingly enough, largely true. There is no generic 'First Class status' - different communities excel in different areas of biotech. Missouri can excel in other areas without being #1 in stem cell research.
- "Reason 17: Amendment 2 is NOT in step with the rest of the world." They go on to say that 27 countries have banned human cloning. But again, they are dishonestly exploiting the multiple definitions of cloning. Some of those countries have passed legislation essentially identical to Amendment 2, which does ban cloning full human beings but allows other research. To use those countries as an argument against amendment 2 is extremely dishonest.
- "Reason 18: Amendment 2 will require your hard earned tax dollars." Recycling again - go back and look at Reason 2. They go on to claim that "private money has been directed to more promising research avenues," which is an outright lie - private money is almost the only source of money currently available for stem cell research in this country. It would be helpful if federally funded labs (meaning, most university research labs) could be involved in stem cell research.
- "Reason 19: Amendment 2 has potential to make some people and corporations very wealthy at Missourians' expense. Researchers and bio-tech corporations stand to make a lot of money from patents - even if no cures ever come from human cloning and embryonic stem cell research." In a sense this is true of all government sponsored research - taxpayers fund research at academic labs, which is then developed into usable drugs and therapies by corporations who earn a profit. The whole thing about patent wealth even if there are no cures - this is a little crazy. Honestly, how many people get wealthy from patents on technologies that don't work?
- "Reason 20: Amendment 2 contains no real internal controls or oversight." Another lie. The internal controls and oversight specified in Amendment 2 are exactly the same as those required for any other research involving human subjects. Universities have to certify to funding agencies that they are compliant with these regulations, every time a researcher submits a grant proposal. There is nothing in Amendment 2 that makes stem cell research exempt from all of the regular oversight that is currently in place for studies like vaccine trials, drug tests, experimental surgeries, etc. Reason 20 does not offer an argument specific to stem cell research.
These talking points are largely based on fear-mongering, dishonest blurring of definitions of cloning, and base appeals against 'elites', the wealthy, and the biotech industry. If you live in Missouri, you should be embarrassed to have a 'Vote NO on Amendment 2' lawn sign (at least ones issued by Missourians Against Human Cloning). If you're not in Missouri, I'm sure you'll see nearly identical arguments at some point in your own state, or if not, again at the federal level once the next Congress is in session.
As a general principle, I think the public (the taxpayers funding scientific research in the US) should be involved discussing what kind of science should be done in our society. Mind you, they shouldn't be judging the scientific, technical merit of specific proposals - that's as absurd as asking someone with no engineering training to evaluate the structural integrity of a proposed bridge design.
But some general input is good. I just submitted a funding proposal to the American Cancer Society. On their review panels, they include non-scientists who are interested in cancer research, and my application includes an explanation in non-technical language of what I'm proposing to do and how it is related to cancer. I am judged partly on how well I can explain my work to a general audience, and, more importantly, on whether I can persuade the non-scientist on the panel that my work is important for cancer research. I think that's a great part of the process.
On the other hand, often in the political arena, people's arguments on science issues are based on pure dishonesty and bad faith. The opposition movement to Missouri's amendment 2 falls under this category; they are lying to scare people into voting against the amendment. In their flyers and on their website, they lecture us about ethics, but they are liars and deserve no credibility on this issue.
These are harsh words, but as you'll see below, almost every reason they give to vote against this bill is either deliberately misleading or based on a flat-out lie.
To see that these people are liars, you have to know first what the amendment says. It's never a good idea to take someone else's word on what a ballot initiative says, so you read it yourself right here. And if you're not a resident of Missouri, you should still be interested, because many other states are wrestling with the same issue right now, not to mention our national debate on the subject.
Here's my summary of what I think is a very reasonable initiative (and really, check me by reading the initiative yourself):
The Main Point: stem cell research and therapies permitted under federal law shall be legal in Missouri. Since very little is actually legal or funded under current federal law (and this amendment isn't providing any money), right now this makes little difference. This amendment is really aimed at a time when, say, Congress could override a Bush veto of a stem cell bill like the one that was recently vetoed. This amendment is just assuring that stem cell research and therapies are treated like any other research. You don't generally see certain mainstream research or therapies banned in certain states, while permitted overall at the federal level.
Important restrictions specified in the amendment:
- no cloning a human being - that is, you can't implant a human embryo in a human uterus, if that embryo was not created with human sperm and egg. A cloned embryo is created without sperm (referred to below as Somatic Cell Nuclear Transfer or SCNT).
- no creating a blastocyst purely for research by fertilization - in other words, embryos taken for stem cells would be left over at in vitro fertilization (IVF) clinics, or created through producing a clone by SCNT (if permitted by federal law).
-no taking cells from a blastocyst after more than 14 days of cell division
-no selling eggs or blastocysts
The amendment also includes typical provisions that already apply to all ongoing research using human subjects or samples; these provisions are redundant, because they are already required by current federal laws and regulations:
- donors of eggs/blastocysts must give informed, voluntary consent
- researchers must comply with normal standards of bioethics, must have Institutional Review Board approval (required at all research universities before someone can work with human subjects or tissue samples), and comply with all other regulations that apply to research with human subjects and samples in general.
This proposal basically deals with all non-fundamentalist ethical issues, and I predict that such an approach will be adopted at the federal level within 4 years (and maybe even during the next Congressional session).
A side note on cloning: Beware of people who try to tell you cloning is cloning is cloning - and that there is no difference, it's all ethically the same. Not true! Scientists use the term cloning to mean different things. Cloning a gene is not the same a cloning a human being. Molecular biologists make copies of genes and put them into a variety of different contexts; they call that cloning. Making a human clone involves implanting a cloned embryo into a uterus, and having the woman eventually deliver the baby. This has not been done yet, and it's unethical because you would probably have hundreds of women suffer hundreds of miscarriages before you actually had a live birth. On top of that, that live child will probably have future health problems (because that's what we've observed in cloned animals). But these human clones would not be soulless robots (the Hollywood-inspired fear expressed by a woman I met on the train the other month) - they would be like the natural clones among us today - identical twins, fully autonomous human beings.
Cloning embryos for stem cell research starts out the same as human cloning (pull the DNA from an egg, put in DNA from an adult cell, and get it to divide without fertilization by sperm - SCNT), but you don't implant the embryo in a uterus. People worry that doing this though is the first step towards human cloning, and what's to stop us from going all the way? Well, the fact that cloning an embryo is the easy part, getting it to develop in the uterus is the really hard part. And also, the ethical issues significantly change once you implant such an embryo in the uterus, as I stated before.
Back to amendment 2: the proposed amendment is facing a disinformation campaign by a dishonest opposition. The "Missourians Against Human Cloning" have put out 20 talking points, each of which is either severely misleading or an outright lie. If you've had enough of this issue, you can stop reading the post here, but for all the gory details, keep reading:
- "Reason 1: Amendment 2 would permanently change Missouri's Constitution." Permanently, until voters adopt an amendment to change this one. This claim is misleading, because it ignores that fact that, unlike what happens at the federal level, state constitutional amendments are a common way of doing business in most states, and the amendments are easy to change. Missouri has four proposed amendments on the ballot this Fall, most dealing with mundane issues like property tax exemptions and pensions for state officials convicted of felonies. There is nothing unusual about putting an issue like stem cell research into a state constitutional amendment.
- "Reason 2: Amendment 2 will use our tax dollars for unethical and unproven research. Despite claims to the contrary, if Amendment 2 passes, Missourians will pay for unethical and unproven research indefinitely." Also dishonest - the amendment provides NO MONEY. Any money for such research would come from the NIH budget, which is unlikely to get an extra raise specifically for stem cell research. If you want to talk about our tax dollars going to something unethical (and, of unproven or disproven effectiveness), let's talk about the Bush administration's torture policy.
- "Reason 3: Amendment 2 would create a constitutional right to devalue human life - it would treat human life as a commodity and raw material for unethical human experimentation by the bio-tech industry." Here's an outright lie - the amendment specifically prohibits treating eggs or embryos as commodities - that is, something to be bought and sold. Beyond that, the claim that stem cell research for disease cures in some vague, unarticulated way devalues human life is weak. Notice the scare language that implicitly evokes Nazi-style research - stem cell research is 'unethical human experimentation'!
- "Reason 4 - Amendment 2 is harmful to women... up to 35% of women who submit to ovarian stimulation experience health consequences, and up to 14% of these are severe." Those numbers are false. The procedure is uncomfortable, but it's not really more risky than an appendectomy. It's done routinely, every day, all over this country. My wife went through many rounds of ovarian stimulation drugs, and went through egg retrieval twice. These drugs have been used for a long time, they have a long track record, and they are safe in the vast majority of cases (no riskier than the anesthesia used in a routine appendectomy). It's extremely common, and I would be money that you know someone who has taken ovarian stimulation drugs, even though you may not be aware of it.
- "Reason 5 - Amendment 2 is unnecessary - adult stem cell research has been proven effective for many illnesses... We need to retain the option to direct our tax dollars to this ethical and promising research." That's false - there are actually few illnesses that adult stem cells work for - anything requiring a bone marrow transplant, and that's just about it. We always have the option to study adult stem cells, and labs who are already doing this work are not suddenly going to drop it once there is money for embryonic stem cell research. Furthermore, the adult stem cell researchers I know personally are some of the most passionate advocates for embryonic stem cell research, even though they have no intention of dropping all of their current work to study embryonic stem cells should funding become available.
- "Reason 6: Amendment 2 protects and promotes human cloning. The proponents want you to believe that Somatic Cell Nuclear Transfer (SCNT) is NOT cloning, but the reality is that every textbook and scientific journal defines SCNT as Cloning." Here they dishonestly try to muddy the waters by exploiting the fact that scientists use the term cloning in different ways - see my earlier note on cloning. The ethical issues relevant to making a live human clone are different from those involving cloning embryos (or SCNT) that will never see the inside of a uterus. You have to articulate each of these ethical cases individually. I can think of reasons why it's wrong to make human clones (health problems in the cloned humans, emotional and physical risks to the women who carry these pregnancies with a high risk of failure), but those reasons don't apply to embryos that are never implanted in a uterus. The Amendment 2 opponents are think they can just get away with screaming CLONING!!! instead of articulating a serious argument against embryo cloning. This is a tactic that repeatedly shows up in this debate.
- "Reason 7 Amendment 2 is an elite initiative" - basically, 95% of the promotional money for this amendment came from one wealthy couple. This talking point is not dishonest, it's just stupid. We'll see how elite this amendment really is when we vote on it next month.
- "Reason 8 - Amendment 2 is immoral. It is ethically wrong to destroy human life regardless of its origin or geography." I'm not sure what geography has to do with anything here, but if Amendment 2 is immoral because it 'destroys human life', then so is the process used for in vitro fertilization and many common forms of birth control. Most citizens don't seem to be outraged over birth control or IVF, so I take it that most people don't buy this ethical reasoning.
- "Reason 9 - Amendment 2 will exploit the disadvantaged" because they will be paid to donate their eggs for research. In reality, this won't exploit the 'disadvantaged' any more than any other research involving human subjects - people who volunteer for such studies are paid a small fee in return for their time, effort, and travel expenses. This is how all new drugs and treatments are tested - if amendment 2 is exploitative, so is essentially all research with human subjects. Many such studies involve procedures much more risky than egg donation.
- "Reason 10: Amendment 2 is wrong. To permanently change our constitution to protect unproven and risky scientific experimentation is wrong." We're only halfway to Reason 20, and already they're recycling their previous reasons (see Reasons 1, 2, and 5). I guess this means that there really aren't 20 independent reasons to vote against Amendment 2.
- "Reason 11: Amendment 2 ignores proven research. Ethical stem cell research provides real hope without cloning or destroying human life..." Now they're recycling Reason 5.
- "Reason 12: Amendment 2 is deceptive - it will PROMOTE not ban human cloning." Recycling Reason 6 here. Like I said above, this is a dishonest and misleading attempt to muddy the various meanings of 'cloning'. Amendment 2 bans cloning human beings, but it does not ban the creation of cloned human embryos that will never be implanted in a uterus. Amendment 2 is clear about it's definitions (again, go read the entire thing yourself); the Missourians Against Human Cloning are the ones being deceptive.
- "Reason 13: Amendment 2 will destroy human life." Amendment 2 could save human lives through the treatments it might facilitate. Unless you think destroying blastocysts is equal to murder, Reason 13 is not true.
- "Reason 14: Amendment 2 will NOT give Missourians more access rights to cures or therapies that they and every other American ALREADY enjoy." This is dishonest - that's not the point of the amendment; the point is to make sure that future therapies permitted elsewhere aren't banned in Missouri.
- "Reason 15: Amendment 2 ignores the success of Adult Stem Cell therapies and cures." Wow, we're recycling Reason 5 for a second time. Amendment 2 does not 'ignore' Adult stem cell research - it doesn't ban it and it doesn't take away research dollars (especially since it doesn't provide any research dollars for embryonic stem cell research). And naturally adult stem cells therapies are farther along than embryonic stem cell therapies - adult stem cell research has been going on longer, and that research hasn't been blocked at the federal level.
- "Reason 16: Amendment 2 will NOT insure First Class status to Missouri's biotech industry." This is, amazingly enough, largely true. There is no generic 'First Class status' - different communities excel in different areas of biotech. Missouri can excel in other areas without being #1 in stem cell research.
- "Reason 17: Amendment 2 is NOT in step with the rest of the world." They go on to say that 27 countries have banned human cloning. But again, they are dishonestly exploiting the multiple definitions of cloning. Some of those countries have passed legislation essentially identical to Amendment 2, which does ban cloning full human beings but allows other research. To use those countries as an argument against amendment 2 is extremely dishonest.
- "Reason 18: Amendment 2 will require your hard earned tax dollars." Recycling again - go back and look at Reason 2. They go on to claim that "private money has been directed to more promising research avenues," which is an outright lie - private money is almost the only source of money currently available for stem cell research in this country. It would be helpful if federally funded labs (meaning, most university research labs) could be involved in stem cell research.
- "Reason 19: Amendment 2 has potential to make some people and corporations very wealthy at Missourians' expense. Researchers and bio-tech corporations stand to make a lot of money from patents - even if no cures ever come from human cloning and embryonic stem cell research." In a sense this is true of all government sponsored research - taxpayers fund research at academic labs, which is then developed into usable drugs and therapies by corporations who earn a profit. The whole thing about patent wealth even if there are no cures - this is a little crazy. Honestly, how many people get wealthy from patents on technologies that don't work?
- "Reason 20: Amendment 2 contains no real internal controls or oversight." Another lie. The internal controls and oversight specified in Amendment 2 are exactly the same as those required for any other research involving human subjects. Universities have to certify to funding agencies that they are compliant with these regulations, every time a researcher submits a grant proposal. There is nothing in Amendment 2 that makes stem cell research exempt from all of the regular oversight that is currently in place for studies like vaccine trials, drug tests, experimental surgeries, etc. Reason 20 does not offer an argument specific to stem cell research.
These talking points are largely based on fear-mongering, dishonest blurring of definitions of cloning, and base appeals against 'elites', the wealthy, and the biotech industry. If you live in Missouri, you should be embarrassed to have a 'Vote NO on Amendment 2' lawn sign (at least ones issued by Missourians Against Human Cloning). If you're not in Missouri, I'm sure you'll see nearly identical arguments at some point in your own state, or if not, again at the federal level once the next Congress is in session.
Tuesday, October 03, 2006
Nobel week kicks off with a well-deserved award for RNAi
It's Nobel week, and I'm enough of a nerd to anxiously look forward to this week every year. Yesterday the Nobel committee annouced that the Medicine/Physiology prize is going to Andrew Fire and Craig Mello. Fire is currently at Stanford, and Mello is at U. Mass in Worcester.
It's great to see the prize go to scientists who are under 50, recognizing work that was done less than a decade ago. When the prize recognizes very old work too often, the Nobel loses much of its excitement and risks losing its relevance to current science. The original Nature paper by Fire, Mello, and their colleagues, came out in Feb. 1998. Since that time, RNAi has become huge in the field of biology - both in terms of its role in the cell, as a powerful genetics tool, and as a potential way to do gene therapy.
RNAi, or RNA interference, is when the expression of a single gene is shut off by the presence of foreign RNA molecules that match part of the sequence of that gene. RNAi occurs in plants, animals, and fungi, and most likely evolved as a defense against viruses and other genomic parasites (such as transposons - they're like viruses that spread through the genome and never leave the cell; we all have a lot of these parasites hanging around in our genomes).
What did Fire and Mello do? A look at their work illustrates how major scientific discoveries don't just come out of thin air - they are based on the groundwork usually laid down by several different research groups. Fire and Mello didn't discover RNAi - scientists had known for years that injecting small RNA molecules into organisms like round worms (such as C. elegans - the organism Fire and Mello work with) could inhibit the expression of single genes. Similar phenomena had been described for both plants and bread mold. (Bread mold - N. crassa - is another weird organism that has been used to study many basic cellular processes.)
People hypothesized that it worked because injected 'anti-sense' RNA bound (or 'hybridized') to the cell's messenger RNA (mRNA) and prevented that mRNA from serving as a template for synthesizing new protein:
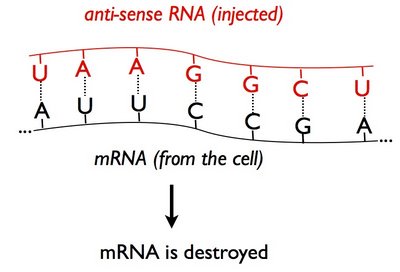
(Recall that the letters A,C,G, and in RNA, U stand for chemical groups that match up with each other - A pairs with U, C pairs with G.)
But there were problems with this idea - it also worked with 'sense' RNA as well, which would not bind to the mRNA molecule. Also, a small number of injected RNA molecules, not nearly enough to bind up all of the mRNA produced by a partuclar gene, could cause RNAi - suggesting that the injected RNA molecules were reused over and over to break down the mRNA (that is, they funtioned catalytically). This effect could even been seen in the next generation of organisms, that had not been injected with foreign RNA.
Fire, Mello and colleagues came up with the brilliant idea that the phenomenon might be caused by double-stranded RNA molecules, which would have been present as contaminants in any injected sample of RNA. Such double-stranded molecules would not work by simply hybridizing to the mRNA (as shown in the figure above). The process would have to work by some other mechanism, which at the time was completely unknown.
So Fire and Mello deliberately prepared sense, anti-sense, and double-stranded RNAs, injected them into worms, and looked to see which RNA molecules would be most effective at knocking down gene expression.
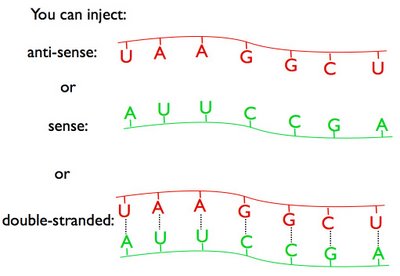
They picked a clever target gene to knock down - unc-22, a gene coding for a muscle protein. When that muscle protein is absent, the worms uncontrollably twitch. So Fire and Mello injected the different RNA molecules into worm embryos, and looked to see which ones resulted in twitching adult worms, indicating that the unc-22 gene had been shut off. Their results conclusively demonstrated that it was the double-stranded RNA molecules, not the individual sense or anti-sense molecules, that caused RNAi.
This paved the way for later discoveries figuring out excatly how this process works, and for making huge collections of these RNAs that can be used to knock down just about any gene you want in model organisms like flies and worms. Until RNAi, you couldn't shut off genes so easily, except in microorganisms like yeast (which is one reason why yeast is so useful for studying basic molecular biology). Now, you can do those things in multi-cellular organisms, and hardly a week goes by without the report of some new discovery made using RNAi. These double-stranded RNA molecules are even being tested as drugs in humans, to shut off aberrant genes in people with certain diseases. This was certainly a timely and well-deserved Nobel prize.
It's great to see the prize go to scientists who are under 50, recognizing work that was done less than a decade ago. When the prize recognizes very old work too often, the Nobel loses much of its excitement and risks losing its relevance to current science. The original Nature paper by Fire, Mello, and their colleagues, came out in Feb. 1998. Since that time, RNAi has become huge in the field of biology - both in terms of its role in the cell, as a powerful genetics tool, and as a potential way to do gene therapy.
RNAi, or RNA interference, is when the expression of a single gene is shut off by the presence of foreign RNA molecules that match part of the sequence of that gene. RNAi occurs in plants, animals, and fungi, and most likely evolved as a defense against viruses and other genomic parasites (such as transposons - they're like viruses that spread through the genome and never leave the cell; we all have a lot of these parasites hanging around in our genomes).
What did Fire and Mello do? A look at their work illustrates how major scientific discoveries don't just come out of thin air - they are based on the groundwork usually laid down by several different research groups. Fire and Mello didn't discover RNAi - scientists had known for years that injecting small RNA molecules into organisms like round worms (such as C. elegans - the organism Fire and Mello work with) could inhibit the expression of single genes. Similar phenomena had been described for both plants and bread mold. (Bread mold - N. crassa - is another weird organism that has been used to study many basic cellular processes.)
People hypothesized that it worked because injected 'anti-sense' RNA bound (or 'hybridized') to the cell's messenger RNA (mRNA) and prevented that mRNA from serving as a template for synthesizing new protein:

(Recall that the letters A,C,G, and in RNA, U stand for chemical groups that match up with each other - A pairs with U, C pairs with G.)
But there were problems with this idea - it also worked with 'sense' RNA as well, which would not bind to the mRNA molecule. Also, a small number of injected RNA molecules, not nearly enough to bind up all of the mRNA produced by a partuclar gene, could cause RNAi - suggesting that the injected RNA molecules were reused over and over to break down the mRNA (that is, they funtioned catalytically). This effect could even been seen in the next generation of organisms, that had not been injected with foreign RNA.
Fire, Mello and colleagues came up with the brilliant idea that the phenomenon might be caused by double-stranded RNA molecules, which would have been present as contaminants in any injected sample of RNA. Such double-stranded molecules would not work by simply hybridizing to the mRNA (as shown in the figure above). The process would have to work by some other mechanism, which at the time was completely unknown.
So Fire and Mello deliberately prepared sense, anti-sense, and double-stranded RNAs, injected them into worms, and looked to see which RNA molecules would be most effective at knocking down gene expression.

They picked a clever target gene to knock down - unc-22, a gene coding for a muscle protein. When that muscle protein is absent, the worms uncontrollably twitch. So Fire and Mello injected the different RNA molecules into worm embryos, and looked to see which ones resulted in twitching adult worms, indicating that the unc-22 gene had been shut off. Their results conclusively demonstrated that it was the double-stranded RNA molecules, not the individual sense or anti-sense molecules, that caused RNAi.
This paved the way for later discoveries figuring out excatly how this process works, and for making huge collections of these RNAs that can be used to knock down just about any gene you want in model organisms like flies and worms. Until RNAi, you couldn't shut off genes so easily, except in microorganisms like yeast (which is one reason why yeast is so useful for studying basic molecular biology). Now, you can do those things in multi-cellular organisms, and hardly a week goes by without the report of some new discovery made using RNAi. These double-stranded RNA molecules are even being tested as drugs in humans, to shut off aberrant genes in people with certain diseases. This was certainly a timely and well-deserved Nobel prize.
Tuesday, September 26, 2006
Tierney weighs in on the Gender Bias Panel
In my last post I discussed the recent National Academies panel on gender bias. In today's NY Times, John Tierney weighs in on the issue. (This one is 'Times Select' - subscription required.)
I'm usually at ideological odds with Tierney, but not quite so much this time. He comes up with a great Onion title for the panel's work:
"This is the kind of science you expect to find in The Onion: 'Academy Forms Committee to Study Gender Discrimination, Bars Men from Participating.' Actually, it did allow a total of one man, Robert Birgeneau of Berkeley, on the 18-member committee, but that was presumably because he was already on record agreeing with the report’s pre-ordained conclusion: academia must stop favoring male scientists and engineers."
Tierney hits some good points (such as how the panel fails to distinguish between bias 30 years ago and bias today). He also gets into the 'innate differences' issue that I avoided in my last post:
"One well-documented difference is the disproportionately large number of boys scoring in the top percentile of the SAT math test. And when you compare boy math whizzes with girl math whizzes, more differences appear. The boys score much higher on the math portion of the SAT than on the verbal, whereas the girls are more balanced — high on the verbal as well as the math.
The girls have more career options, and they have different priorities than the boys, as the psychologists David Lubinski and Camilla Persson Benbow have demonstrated by tracking students with the exceptional mathematical ability to become top-flight researchers in science and engineering."
I've got two things to note about this:
- In addition to a disproportionate number of boys in the top math SAT percentile, there is also a disproportionate number of boys in the bottom percentile. In other words, the average score is not so different among boys and girls, but there are more boys on both ends of the curve.
- This difference in the top percentile in SAT math may be relevant to differences in some sciences like theoretical physics or pure math (I have no idea), but, as I hinted at in my last post, I think it's irrelevant in many other fields like biology. Biology has some very quantitative aspects, but the people I know who are outstanding in those areas aren't necessarily the ones who were 'math whizzes' on the SAT. Good biologists need a variety of different skills, not just the ones that lead to a top SAT math score. Also, in biology, rigorous verbal reasoning skills play a much larger role, and recall that girls are overrepresented in the top percentiles of the verbal SAT. So are women innately better biologists?
One statement in Tierney's article especially bugs me- he lumps biology with psychology as a 'soft science.' This idea that men are better at 'hard' sciences and women are better at 'soft' sciences is just crap. Biology is just as much a hard (meaning, roughly, quantitative and experimentally rigorous) science as any other of the natural sciences - geology, astronomy, and yes, physics and chemistry. Biology (evolution, molecular biology, genetics, ecology, etc.) has much more in common with astronomy or geology than with psychology.
UPDATE: If you can access this (subscription required), this Nature Neuroscience Editorial has some interesting references to the primary literature and comments on the differences in SAT scores. They note that the score differences (more boys at the tail ends of the curve) don't occur in all Western countries, suggesting that the effect may very well be cultural. Who knows? It seems like the debate is often just stuck at the level of "Women are less able!" "No they're not, men are just hopelessly predjudiced!" As I made clear in my last post, I think there are other answers.
[This post was edited a few times for clarity.]
I'm usually at ideological odds with Tierney, but not quite so much this time. He comes up with a great Onion title for the panel's work:
"This is the kind of science you expect to find in The Onion: 'Academy Forms Committee to Study Gender Discrimination, Bars Men from Participating.' Actually, it did allow a total of one man, Robert Birgeneau of Berkeley, on the 18-member committee, but that was presumably because he was already on record agreeing with the report’s pre-ordained conclusion: academia must stop favoring male scientists and engineers."
Tierney hits some good points (such as how the panel fails to distinguish between bias 30 years ago and bias today). He also gets into the 'innate differences' issue that I avoided in my last post:
"One well-documented difference is the disproportionately large number of boys scoring in the top percentile of the SAT math test. And when you compare boy math whizzes with girl math whizzes, more differences appear. The boys score much higher on the math portion of the SAT than on the verbal, whereas the girls are more balanced — high on the verbal as well as the math.
The girls have more career options, and they have different priorities than the boys, as the psychologists David Lubinski and Camilla Persson Benbow have demonstrated by tracking students with the exceptional mathematical ability to become top-flight researchers in science and engineering."
I've got two things to note about this:
- In addition to a disproportionate number of boys in the top math SAT percentile, there is also a disproportionate number of boys in the bottom percentile. In other words, the average score is not so different among boys and girls, but there are more boys on both ends of the curve.
- This difference in the top percentile in SAT math may be relevant to differences in some sciences like theoretical physics or pure math (I have no idea), but, as I hinted at in my last post, I think it's irrelevant in many other fields like biology. Biology has some very quantitative aspects, but the people I know who are outstanding in those areas aren't necessarily the ones who were 'math whizzes' on the SAT. Good biologists need a variety of different skills, not just the ones that lead to a top SAT math score. Also, in biology, rigorous verbal reasoning skills play a much larger role, and recall that girls are overrepresented in the top percentiles of the verbal SAT. So are women innately better biologists?
One statement in Tierney's article especially bugs me- he lumps biology with psychology as a 'soft science.' This idea that men are better at 'hard' sciences and women are better at 'soft' sciences is just crap. Biology is just as much a hard (meaning, roughly, quantitative and experimentally rigorous) science as any other of the natural sciences - geology, astronomy, and yes, physics and chemistry. Biology (evolution, molecular biology, genetics, ecology, etc.) has much more in common with astronomy or geology than with psychology.
UPDATE: If you can access this (subscription required), this Nature Neuroscience Editorial has some interesting references to the primary literature and comments on the differences in SAT scores. They note that the score differences (more boys at the tail ends of the curve) don't occur in all Western countries, suggesting that the effect may very well be cultural. Who knows? It seems like the debate is often just stuck at the level of "Women are less able!" "No they're not, men are just hopelessly predjudiced!" As I made clear in my last post, I think there are other answers.
[This post was edited a few times for clarity.]
Monday, September 25, 2006
The long road to a career in academic biology...
What's a postdoc? 90% of the time, that's the next question I'm asked after people ask me what I do for a living. Or worse, people who know I spent 5 and a half years in grad school will say "wow, I can't believe you're still a student!" It's clear that most people outside the scientific community don't know what postdocs are - Students? Interns? Trainees?
People should know what postdocs are - at very least so that they don't call them students! More importantly, the public should know who is actually doing the vast majority of the experiments and fieldwork that get published in hundreds of scientific journals every week: postdoctoral fellows and grad students. They are the people who pick up the test tubes and pipettes, go the lab bench, and run the experiments. That's not to say that the professor just sits back and gets all the credit - professors are generally full intellectual participants in the research, and they're usually the ones who came up with the main idea for the research project. However, the point here is that grad students and postdocs aren't simply trainees learning their trade; they are practicing scientists who produce valuable, tangible work. Unlike undergraduate education, this is on-the-job training much like any other career field.
Postdocs are no longer students - they have their PhDs, and they're not attending classes, taking tests, or writing a thesis. If a graduate student is an apprentice, a postdoc is a journeyman - a credentialed, capable scientist who is gaining more experience under the guidance of a senior scientist. A postdoctoral position is a chance to learn some new skills and work more independently without having to jump through the administrative hoops of grad school. The research you do as a postdoc lays the foundation for what you'll do in your own lab as a professor.
I still seem like a student to my friends in other careers because my job as a postdoc is only temporary (3-5 years), and I still make well below the median US income. (Starting postdoc salaries range from $30k to ~$40k, and rise to $40-$45k after 2 years of experience.) This is why the career path of an academic scientist seems so long - you hardly earn anything and don't settle down until you're in your mid- to late 30's, even though you have a doctorate and you do demanding, highly technical work. You spend 5+ years as a grad student with a 22k annual salary, no retirement benefits, and bare-bones health insurance. Your reward for earning a PhD? A temporary job with a low salary, no retirement benefits, and if you're lucky, decent health insurance. All this, while your friends who went to law school finished their degrees years ago, got paid $50-60k during a brief clerkship, and are now raking in more money than you'll make as a tenured scientist.
On the other hand, if you work with a good mentor, you wouldn't trade this job for any other. In spite of the unsustainable pay level (you'll never retire or send your kids to college on it), as a science postdoc you get to continually drive your intellect and creativity, you get to play with fun, high-tech toys, and you have a degree of independence that almost rivals that of a freelance writer.
People should know what postdocs are - at very least so that they don't call them students! More importantly, the public should know who is actually doing the vast majority of the experiments and fieldwork that get published in hundreds of scientific journals every week: postdoctoral fellows and grad students. They are the people who pick up the test tubes and pipettes, go the lab bench, and run the experiments. That's not to say that the professor just sits back and gets all the credit - professors are generally full intellectual participants in the research, and they're usually the ones who came up with the main idea for the research project. However, the point here is that grad students and postdocs aren't simply trainees learning their trade; they are practicing scientists who produce valuable, tangible work. Unlike undergraduate education, this is on-the-job training much like any other career field.
Postdocs are no longer students - they have their PhDs, and they're not attending classes, taking tests, or writing a thesis. If a graduate student is an apprentice, a postdoc is a journeyman - a credentialed, capable scientist who is gaining more experience under the guidance of a senior scientist. A postdoctoral position is a chance to learn some new skills and work more independently without having to jump through the administrative hoops of grad school. The research you do as a postdoc lays the foundation for what you'll do in your own lab as a professor.
I still seem like a student to my friends in other careers because my job as a postdoc is only temporary (3-5 years), and I still make well below the median US income. (Starting postdoc salaries range from $30k to ~$40k, and rise to $40-$45k after 2 years of experience.) This is why the career path of an academic scientist seems so long - you hardly earn anything and don't settle down until you're in your mid- to late 30's, even though you have a doctorate and you do demanding, highly technical work. You spend 5+ years as a grad student with a 22k annual salary, no retirement benefits, and bare-bones health insurance. Your reward for earning a PhD? A temporary job with a low salary, no retirement benefits, and if you're lucky, decent health insurance. All this, while your friends who went to law school finished their degrees years ago, got paid $50-60k during a brief clerkship, and are now raking in more money than you'll make as a tenured scientist.
On the other hand, if you work with a good mentor, you wouldn't trade this job for any other. In spite of the unsustainable pay level (you'll never retire or send your kids to college on it), as a science postdoc you get to continually drive your intellect and creativity, you get to play with fun, high-tech toys, and you have a degree of independence that almost rivals that of a freelance writer.
Monday, September 18, 2006
National Academy panel tries to tell us how to keep more women in science:
The NY Times has published an article titled Institutions Hinder Female Academics, Panel Says. (You can check out the full report and the panel's press release.) The panel in question was convened by the National Academy of Sciences to look at how women are faring in academic science and technology careers. Unfortunately, I think this whole issue is tainted by ideology, resulting in often illogical arguments. OK I confess, I'm male and I'm white, but give me a chance here.
Some of the claims made by this panel (and by others in this debate) are absurd, and their suggestions threaten to pile more ridiculous bureaucracy on already overburdened academic administrators. But let me start with some postive comments first:
The chair of the panel, Donna E. Shalala, said "The United States should enhance its talent pool by making the most of its entire population." Absolutely! Women should not be discriminated against, period. And yes, women were severely discriminated against in academic science not too long ago. As far as 'innate differences' in generaly ability go, in my field of biology it seems pretty obvious that there aren't any - plenty of women are first-rate biologists. (There's more on this 'innate ablity' issue, but that's for another post, another day.)
In fact, because their are so many first-rate women in my field, I have a hard time believing that there is "unconscious but pervasive bias, and 'arbitrary and subjective' evaluation processes..." The men in the departments I have been in, from grad students to full professors, work with these outstanding women every day - there is simply no way they could believe that these women were somehow not as suitable for science. The claim that there is some unconscious cultural bias against women is flat out wrong. What kind of serious, specific evidence for unconscious bias does this panel have? They have none - at least none that can't be better explained by other causes that I'll get to below.
I'm struggling here to convey how far-fetched these claims of patronization or bias are when comapred against the every-day reality that I work in. It's just as absurd as claiming there is a bias against Jews in science, that's the best I can put it - successful women are so pervasive in my field, that to say male scientists somehow look down on them is just plain insulting. On top of that, academic scientists tend to be fairly liberal and progressively minded about racial and gender equality. These absurd claims of bias just don't fit the culture.
Many of the recommendations by this panel are outrageous. Look at this:
"> Measures of success underlying performance-evaluation systems are often arbitrary and frequently applied in ways that place women at a disadvantage. "Assertiveness," for example, may be viewed as a socially unacceptable trait for women but suitable for men."
These people really think that tenure committees members are thinking "Well, she does great work, she has nice legs, but her assertiveness is so unseemly in a woman,"??? Scientists love being with other assertive people, men or women. Asking hard questions is a good thing - it's banged into your head from your first week in graduate school. When people ask me hard questions after I give a talk, I take it as a compliment - it means they are interested in my work, and that I explained it clearly enough for the audience to follow. When I get no questions, I'm disappointed. Most scientists, men and women, feel the same way. The top women in my field, the ones who have been promoted by those misogynist tenure committees, are very assertive. As they should be. And they're not viewed as 'more masculine' (as I've heard some partisans claim), 'underfeminized' or somehow abnormal of their gender. And there's not some checkbox for 'assertiveness' in performance evaluations, as the panel seems to imply.
Another recommendation by this panel is that "Federal funding agencies and foundations, in collaboration with professional and scientific societies, should hold mandatory national meetings to educate university department chairs, agency program officers, and members of review panels on ways to minimize the effects of gender bias in performance evaluations." I can't think of a bigger waste of time than to have department chairs and reviewers fly out to Washington every so often and be indoctrinated in how not to be biased. These people know how to identify good science, and as I said above, I think the claims of 'unconscious bias' are insulting and absurd.
Muddled up in all of this insulting smearing of male scientists - men who are well educated and culturally progressive - is a discussion of the real problem:
"Also, structural constraints and expectations built into academic institutions assume that faculty members have substantial support from their spouses. Anyone lacking the career and family support traditionally provided by a "wife" is at a serious disadvantage in academe, evidence shows. Today about 90 percent of the spouses of women science and engineering faculty are employed full time. For the spouses of male faculty, it is nearly half. "
This is really the heart of it. As an academic scientist, no one can really fill in for you when you have to take time off for your family. Sure, other people can teach your classes, but nobody can run your lab for you. Nobody can come up with your ideas for you, design your experiments for you, recruit graduate students and postdocs for you, or write your papers and grant proposals for you. If you have to take maternity leave, or go part-time to care for kids, it's hard to keep up, and the progress in your lab slows down. The university may give you all the paid time off you need, but it still won't help when it comes to keeping a spot at the top of your field.
The situation is exacerbated by the long, poverty-ridden training period in a scientist's career. The financial pressure to stay single or childless is strong during this period. It's hard to take time off to have kids when you're a grad student with a poverty-level salary, bad health insurance, and no money going towards a retirement fund. The situation gets a little better when you're a postdoc, but not much. And all of this lasts for 10 years or even more. That financial pressure goes away to some degree when you land a tenure-track job, but then you're faced with a choice - take time off (or go part time) for kids, or drive your career forward. The pressure is there for men too (I've felt it, believe me), but as the report points out, men more frequently have spousal support at home. And even if women did have equal spousal support, child-bearing and raising still have a greater time/physical/emotional impact on women. The decision between a career or full-time parenthood will always be a hard one for women - the pressures of biology are strong enough that there will always be more women than men who drop out of the workforce to become full-time parents, no matter how supportive our institutions are.
Many of us, if not most of us, really do want to become parents at some point. Deciding to never have kids is a huge decision. Equally huge, is deciding whether to stay home or go back to a career. It's hard to decide whether to send your kids to day care or after-school care, and only see them before 8:30 am and after 6 at night (especially if both spouses have careers). It's hard to juggle the time pressures. Most of your free time vaporizes when you have kids, and a career sucks away whatever is left. And no free time just sucks.
No amount of mandatory sensitivity meetings in Washington will cure this. What we really need is room for people to slow things down if they need it. Deciding how to balance career and family is an intensely personal one, and universities can give their scientists some breathing room. Here a few suggestions:
- More universities should allow you to 'stop the tenure clock' if you need it for a year or so to devote time to family. The tenure decision can be thus delayed by a year.
- Universities and funding agencies should make money available for women to restart their labs after time away. Taking time off means you miss several grant cycles and end up with no money to run your lab. Not having grant funding is a very big deal when it comes to getting tenure. Removing that pressure for awhile after maternity leave would help immensely. So would having institutional support for women who want time off when the time is ticking away on a currently funded grant. Grant review committees can have money earmarked for this too.
- Make better health and financial benefits available to women scientists while they are still in training. While it's probably impossible to not lose some ground on your thesis over a break, (if you take a year off, you can't really go back and start at the same place - the field will have moved on, and you'll need to pick a more relevant research question), some planning and support from thesis advisors and departments would help a lot. Outside finanical support is important too - a thesis advisor, already on a tight budget, can't afford to support someone who is not working in the lab.
It's tough - everyone's competing for precious tenure-track slots, universities put tremendous pressure on their academic scientists to bring in grant money, and individual scientific fields move fast. I often wish science in general would slow down - James Watson famously had a lot of spare time on his hands when he and Crick were working on the struture of DNA. Adding federal regulations will just make things worse by sucking up more of people's time (and money!!). What we really need is more money to support women who want to take time off or slow down. If individual scientists, department heads, and university administrators weren't under such financial pressure, we could keep more outstanding women scientists in the top levels of the profession.
Some of the claims made by this panel (and by others in this debate) are absurd, and their suggestions threaten to pile more ridiculous bureaucracy on already overburdened academic administrators. But let me start with some postive comments first:
The chair of the panel, Donna E. Shalala, said "The United States should enhance its talent pool by making the most of its entire population." Absolutely! Women should not be discriminated against, period. And yes, women were severely discriminated against in academic science not too long ago. As far as 'innate differences' in generaly ability go, in my field of biology it seems pretty obvious that there aren't any - plenty of women are first-rate biologists. (There's more on this 'innate ablity' issue, but that's for another post, another day.)
In fact, because their are so many first-rate women in my field, I have a hard time believing that there is "unconscious but pervasive bias, and 'arbitrary and subjective' evaluation processes..." The men in the departments I have been in, from grad students to full professors, work with these outstanding women every day - there is simply no way they could believe that these women were somehow not as suitable for science. The claim that there is some unconscious cultural bias against women is flat out wrong. What kind of serious, specific evidence for unconscious bias does this panel have? They have none - at least none that can't be better explained by other causes that I'll get to below.
I'm struggling here to convey how far-fetched these claims of patronization or bias are when comapred against the every-day reality that I work in. It's just as absurd as claiming there is a bias against Jews in science, that's the best I can put it - successful women are so pervasive in my field, that to say male scientists somehow look down on them is just plain insulting. On top of that, academic scientists tend to be fairly liberal and progressively minded about racial and gender equality. These absurd claims of bias just don't fit the culture.
Many of the recommendations by this panel are outrageous. Look at this:
"> Measures of success underlying performance-evaluation systems are often arbitrary and frequently applied in ways that place women at a disadvantage. "Assertiveness," for example, may be viewed as a socially unacceptable trait for women but suitable for men."
These people really think that tenure committees members are thinking "Well, she does great work, she has nice legs, but her assertiveness is so unseemly in a woman,"??? Scientists love being with other assertive people, men or women. Asking hard questions is a good thing - it's banged into your head from your first week in graduate school. When people ask me hard questions after I give a talk, I take it as a compliment - it means they are interested in my work, and that I explained it clearly enough for the audience to follow. When I get no questions, I'm disappointed. Most scientists, men and women, feel the same way. The top women in my field, the ones who have been promoted by those misogynist tenure committees, are very assertive. As they should be. And they're not viewed as 'more masculine' (as I've heard some partisans claim), 'underfeminized' or somehow abnormal of their gender. And there's not some checkbox for 'assertiveness' in performance evaluations, as the panel seems to imply.
Another recommendation by this panel is that "Federal funding agencies and foundations, in collaboration with professional and scientific societies, should hold mandatory national meetings to educate university department chairs, agency program officers, and members of review panels on ways to minimize the effects of gender bias in performance evaluations." I can't think of a bigger waste of time than to have department chairs and reviewers fly out to Washington every so often and be indoctrinated in how not to be biased. These people know how to identify good science, and as I said above, I think the claims of 'unconscious bias' are insulting and absurd.
Muddled up in all of this insulting smearing of male scientists - men who are well educated and culturally progressive - is a discussion of the real problem:
"Also, structural constraints and expectations built into academic institutions assume that faculty members have substantial support from their spouses. Anyone lacking the career and family support traditionally provided by a "wife" is at a serious disadvantage in academe, evidence shows. Today about 90 percent of the spouses of women science and engineering faculty are employed full time. For the spouses of male faculty, it is nearly half. "
This is really the heart of it. As an academic scientist, no one can really fill in for you when you have to take time off for your family. Sure, other people can teach your classes, but nobody can run your lab for you. Nobody can come up with your ideas for you, design your experiments for you, recruit graduate students and postdocs for you, or write your papers and grant proposals for you. If you have to take maternity leave, or go part-time to care for kids, it's hard to keep up, and the progress in your lab slows down. The university may give you all the paid time off you need, but it still won't help when it comes to keeping a spot at the top of your field.
The situation is exacerbated by the long, poverty-ridden training period in a scientist's career. The financial pressure to stay single or childless is strong during this period. It's hard to take time off to have kids when you're a grad student with a poverty-level salary, bad health insurance, and no money going towards a retirement fund. The situation gets a little better when you're a postdoc, but not much. And all of this lasts for 10 years or even more. That financial pressure goes away to some degree when you land a tenure-track job, but then you're faced with a choice - take time off (or go part time) for kids, or drive your career forward. The pressure is there for men too (I've felt it, believe me), but as the report points out, men more frequently have spousal support at home. And even if women did have equal spousal support, child-bearing and raising still have a greater time/physical/emotional impact on women. The decision between a career or full-time parenthood will always be a hard one for women - the pressures of biology are strong enough that there will always be more women than men who drop out of the workforce to become full-time parents, no matter how supportive our institutions are.
Many of us, if not most of us, really do want to become parents at some point. Deciding to never have kids is a huge decision. Equally huge, is deciding whether to stay home or go back to a career. It's hard to decide whether to send your kids to day care or after-school care, and only see them before 8:30 am and after 6 at night (especially if both spouses have careers). It's hard to juggle the time pressures. Most of your free time vaporizes when you have kids, and a career sucks away whatever is left. And no free time just sucks.
No amount of mandatory sensitivity meetings in Washington will cure this. What we really need is room for people to slow things down if they need it. Deciding how to balance career and family is an intensely personal one, and universities can give their scientists some breathing room. Here a few suggestions:
- More universities should allow you to 'stop the tenure clock' if you need it for a year or so to devote time to family. The tenure decision can be thus delayed by a year.
- Universities and funding agencies should make money available for women to restart their labs after time away. Taking time off means you miss several grant cycles and end up with no money to run your lab. Not having grant funding is a very big deal when it comes to getting tenure. Removing that pressure for awhile after maternity leave would help immensely. So would having institutional support for women who want time off when the time is ticking away on a currently funded grant. Grant review committees can have money earmarked for this too.
- Make better health and financial benefits available to women scientists while they are still in training. While it's probably impossible to not lose some ground on your thesis over a break, (if you take a year off, you can't really go back and start at the same place - the field will have moved on, and you'll need to pick a more relevant research question), some planning and support from thesis advisors and departments would help a lot. Outside finanical support is important too - a thesis advisor, already on a tight budget, can't afford to support someone who is not working in the lab.
It's tough - everyone's competing for precious tenure-track slots, universities put tremendous pressure on their academic scientists to bring in grant money, and individual scientific fields move fast. I often wish science in general would slow down - James Watson famously had a lot of spare time on his hands when he and Crick were working on the struture of DNA. Adding federal regulations will just make things worse by sucking up more of people's time (and money!!). What we really need is more money to support women who want to take time off or slow down. If individual scientists, department heads, and university administrators weren't under such financial pressure, we could keep more outstanding women scientists in the top levels of the profession.
Subscribe to:
Posts (Atom)



